Abstract
EcoCyc (http://EcoCyc.org) is a comprehensive model organism database for Escherichia coli K-12 MG1655. From the scientific literature, EcoCyc captures the functions of individual E. coli gene products; their regulation at the transcriptional, post-transcriptional and protein level; and their organization into operons, complexes and pathways. EcoCyc users can search and browse the information in multiple ways. Recent improvements to the EcoCyc Web interface include combined gene/protein pages and a Regulation Summary Diagram displaying a graphical overview of all known regulatory inputs to gene expression and protein activity. The graphical representation of signal transduction pathways has been updated, and the cellular and regulatory overviews were enhanced with new functionality. A specialized undergraduate teaching resource using EcoCyc is being developed.
OVERVIEW OF EcoCyc
The EcoCyc user community is diverse: In addition to its use by Escherichia coli biologists, EcoCyc is used by metabolic engineers, by computational biologists and by university educators. Recent enhancements, described below, address user requirements. Our standard literature-based curation practices ensure that the most important findings on E. coli biology are added to EcoCyc in a timely manner.
Because E. coli is the most thoroughly studied bacterium, advances in the understanding of E. coli biology, including the functions of all its genes, are not only of interest to E. coli researchers, but also are helpful for advancing our understanding of less well-studied organisms. Comparative tools are available for the more than 1000 (and growing) databases available in the BioCyc collection (http://biocyc.org), of which EcoCyc is a part. BioCyc tools enable users to navigate the E. coli genome beginning with an orthologous gene in their organism of interest, and to then quickly explore the known functions of the corresponding E. coli genes. High-throughput data can be displayed and analyzed in a larger context—that of the metabolic network, the transcriptional regulatory network, or the entire genome.
UPDATE ON EcoCyc DATA
A team of curators performs curation of the E. coli literature on an ongoing basis. Based on regular PubMed searches, more than 1800 papers with some relevance to the biology of the E. coli laboratory strains are indexed in PubMed every year. Given limited resources, curation of this large amount of literature must be prioritized. At the same time, older entries in EcoCyc must be updated with more recent literature and new data types, such as Gene Ontology (GO) terms. We place the highest curation priority on publications that elucidate previously unknown gene functions and on publications that substantially advance our knowledge of E. coli biology. EcoCyc version 14.5 (September 2010) cites 20 284 publications, and users of the EcoCyc Web site can search the full text of 27 500 publications via Textpresso (1). Table 1 summarizes the current content of EcoCyc.
Table 1.
Overview of the current content of EcoCyc
| Data Type | Number |
|---|---|
| Genes | 4489 |
| Gene products covered by a mini-review | 3666 |
| Enzymes | 1450 |
| Metabolic reactions | 1446 |
| Compounds | 2105 |
| Transporters | 252 |
| Transport reactions | 292 |
| Transported substrates | 207 |
| Transcription factors | 175 |
| Transcription units | 3409 |
| With experimental evidence | 1043 |
| Regulatory interactions | 5345 |
| Transcription initiation | 2746 |
| Transcription attenuation | 18 |
| Enzyme modulation | 2473 |
| Other | 108 |
| Literature citations | 20 284 |
Because we prioritize new functional identifications and significant advances, the data from many publications are not immediately added to EcoCyc. However, our curation strategy includes systems-level updates on the functions of all gene products. These updates include literature searches for a set of genes (for example, all gene products involved in a metabolic pathway), and their curation will be updated with new and older literature and new data types, such as GO terms. Therefore, a significant number of older publications are later cited in EcoCyc. Abstracts and, if freely available, the full text of many E. coli publications, regardless of whether they have been cited in EcoCyc, are added to the Textpresso corpus and can be searched by EcoCyc users.
Update of transcriptional regulation data
EcoCyc knowledge of the transcriptional regulatory network is obtained from the literature, and is continually updated by the RegulonDB biocurator team (2). EcoCyc and RegulonDB releases are synchronized. In addition to our regular curation of knowledge from the literature, we have initiated the annotation of objects from high-throughput methodologies and computational predictions. Curation related to the regulation of transcription initiation is maintained with a delay of one or two months for each release.
Table 2 summarizes the information related to transcriptional regulation published in version 14.5, released in late September 2010. The number of objects has increased considerably since version 12.5; the growth is reported in column 3.
Table 2.
Summary of transcriptional regulation data in EcoCyc
| Data type | Number | Number added since version 12.5 |
|---|---|---|
| Transcription units | 3409 | 53 |
| Promoters | 1878 | 124 |
| Terminators | 239 | 53 |
| Transcription factors | 175 | 12 |
| Transcription factor binding sites | 1940 | 310 |
| Regulatory interactions | 2697 | 355 |
| Small molecule effectors | 77 | 7 |
EcoCyc includes 175 transcription factors (TFs) with at least one experimentally characterized binding site or interaction, belonging to 33 evolutionary families of TFs. Curators author mini-review summaries for TFs and transcription units (TUs) that cover topics such as the regulatory mechanism, growth conditions in which regulation occurs, the cellular processes in which the regulated genes are involved, and a discussion of why those processes might be regulated together.
The 175 TFs are grouped as follows: Seven TFs are considered to be global regulators (ArcA, CRP, FIS, FNR, HNS, IHF, Lrp); 21 response regulators belong to two-component systems; 42 TFs are included in the transport and catabolism of carbohydrates; 17 TFs are related to processes such as transport, biosynthesis and catabolism of the amino acids; 13 TFs are involved in transport and metabolism of different nitrogen sources; and eight TFs are classified as metallo-regulators. Note that individual TFs can be involved in more than one function. The rest of the TFs are considered to be local regulators that control the transcription of genes involved in different cellular processes and functional classes.
Size correction of TF-binding sites (TFBSs)
Most TFs bind to small sequence motifs (7–25 nt) with different symmetries (inverted repeats, direct repeats or asymmetrical sequences) with a variable-size spacer sequence between them. The footprints of a few TFs are longer, from 35 to 60 bp. In general, these long regions do not contain conserved binding sites. Therefore, the binding motif has not been well characterized, even if these interactions are supported by strong evidence, such as: footprinting assay, electrophoretic mobility shift assay or mutations in the potential binding site. We have focused on these TFBSs, and have corrected and relocated several of them. We curated and relocated the binding sites for CytR, OxyR, FhlA based on computational tools [RSAT (3)] that support the identification of overrepresented motifs in regulatory regions. The identification of consensus sequences is based on alignments of these upstream regions, performed by a curator, and on evidence obtained from the literature, including the similarity to the consensus sequence, data from footprinting assays, electrophoretic mobility shift assay, mutational analysis, computational analysis of these sequences and profiling of dependent gene expression.
For example, CytR binding sites were considered 60-bp long based on footprinting experiments. However, now the CytR-binding sites are described as octamer repeats (GTTGCATT) in direct or inverted orientation and preferably separated by 2 bp (4,5). With this information and based on computational analyses, we showed that the binding sites for CytR are overrepresented in the upstream region of the regulated genes. In a similar way, we have relocated, reassigned and corrected binding sites based on shorter binding motifs, for the Ada, YiaJ, NhaR and CaiF TFs.
GO terms
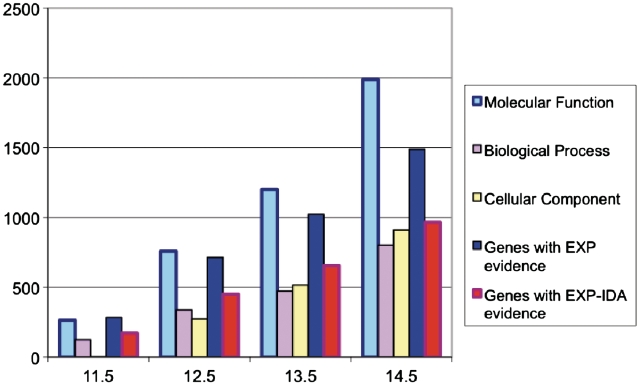
EcoCyc now contains assignments of GO (6,7) terms to a large number of E. coli genes. GO terms are assigned manually by EcoCyc curators during literature curation, and are imported twice a year from UniProt (which contains both computationally predicted and manually curated GO terms) and EcoliWiki. The total number of GO term annotations in EcoCyc is 43 288. Figure 1 shows the growth over time of experimental GO term annotations within EcoCyc. These data provide a lower bound on the number of E. coli genes with experimentally determined functions—a lower bound because GO term curation has been ongoing for a limited time and many well-characterized genes have not had their GO terms manually curated. Note also that many of the GO annotations with experimental evidence reflect approximate functional assignments.
Figure 1.
Growth of experimental GO term annotations in EcoCyc over time. Data is grouped by database version, beginning with release 11.5 (August 2007) and ending with release 14.5 (September 2010). The first three bars in each group report the number of assignments of a GO term from each of the three primary GO categories with an EXP_IDA (inferred by direct assay) evidence code. In contrast, the last two bars report the number of genes that have at least one GO term annotation with an EXP or an EXP-IDA evidence code. EXP includes the EXP-IDA evidence codes as well as others such as EXP-IMP (inferred by mutant phenotype).
Among the uses of GO terms in EcoCyc are retrieval of groups of genes that share a common biological process or molecular function (example: quick search for ‘cell division’, then click GO:0051301—cell division, for a list of all EcoCyc genes annotated to this term). GO terms can also be used in enrichment analysis of gene expression experiments, described below.
Protein features
Protein features are also both manually entered by EcoCyc curators, and are imported from UniProt before every EcoCyc release. UniProt features are derived from both experimental data and computational predictions. EcoCyc v14.5 contains 20 108 features, including enzyme active sites, phosphorylation sites and metal-ion binding sites. When protein features are available for a given protein, those features are displayed toward the bottom of the gene/protein page as annotations on the sequence (e.g. HypB). When importing both GO terms and protein features from UniProt, computationally predicted data are ignored when the same term or feature is present in EcoCyc with experimental evidence.
Protonation and reaction balancing
A barrier to the generation of flux-balance models from EcoCyc has been the fact that EcoCyc metabolites were inconsistently protonated, resulting in reactions that were not balanced for protons (although reactions have been balanced for all other elements for many years). Unbalanced reactions can result in the creation or disappearance of mass in flux-balance models, interfering with the proper functioning of such models. Therefore, we now computationally reprotonate all metabolites in EcoCyc [and MetaCyc (8)] using the Marvin program (ChemAxon) before every release. We also computationally balance every reaction for protons by adding protons to the appropriate side of the reaction.
WEB INTERFACE UPDATES
The EcoCyc web site has undergone the first two phases of a three-phase redesign in collaboration with a user-interface design group at SRI. Phase I consisted of a host of small usability improvements. In Phase II, we introduced a new menu bar, an expanded set of search tools and a new mechanism for selecting which of the hundreds of available BioCyc genomes the user wants to query (click ‘change’ under the Quick Search area at the top right of each page). In Phase III, we will improve the presentation of data content within each page.
EcoCyc now provides multiple search tools that enable searching for specific types of information according to different criteria:
The Quick Search function searches the names of multiple EcoCyc object types (genes, pathways, metabolites, etc.) for terms entered by the user.
Object-Specific searches (such as Search → Genes/Proteins/RNAs) enable the user to define multiple-criteria searches, and then return all objects that match those criteria. For example, the user can search for all genes within a specified base-pair region of the genome, with a specified sequence length and a specified cellular location and that are annotated with specified GO terms.
The Advanced Search function allows the user to specify criteria that span multiple object types (e.g. find all reactions catalyzed by an enzyme with specified properties and containing substrates with specified properties).
Full-Text searches (Search → Full-Text Articles) are implemented using Textpresso (1), which makes the full text of a corpus of 27 500 E. coli articles searchable using keyword and ontology-based searches. For example, the user can search for articles in which the words ‘attenuation’ and ‘tryptophan’ occur in the same sentence. Additional search tools are described at http://biocyc.org/searchhelp.shtml.
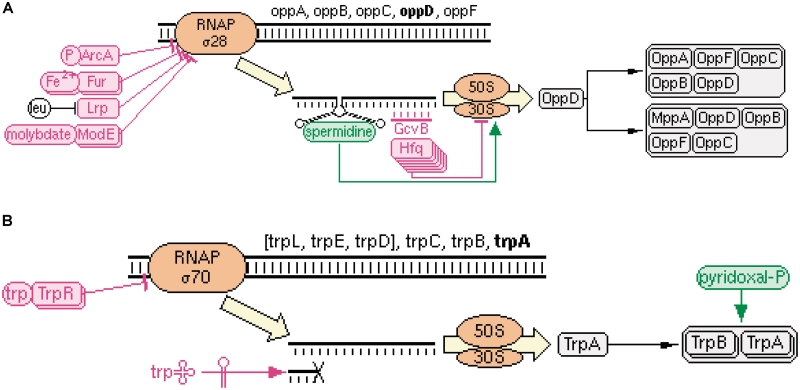
The previously separate EcoCyc web pages describing genes and proteins (or RNA gene products) have been merged to produce a single, integrated page. This merging reduces confusion and unnecessary switching among pages, because it was formerly unclear which kinds of information were found on which pages. A new diagram near the top of every gene page summarizes all the different regulatory inputs on the gene and its product. This diagram includes all factors that regulate transcription initiation, attenuation and translation; the complexes in which the gene product is a component; the post-translational modifications of the protein; and the factors that modulate enzyme activity. Two example diagrams are shown in Figure 2.
Figure 2.
The Regulation Summary Diagram. Colors indicate the overall effect on the production of the functional protein. (A) Regulation Summary Diagram for the oppD gene. Transcription initiation is influenced by a variety of TFs, which are themselves regulated by small molecules. The operon containing oppD is transcribed by RNA polymerase containing the alternative sigma factor Sigma 28. Translation is regulated by a small molecule, spermidine, the small RNA GcvB, and Hfq. The OppD protein can be part of two different heteromultimeric complexes. (B) Regulation Summary Diagram for the trpA gene. Transcription initiation is regulated by TrpR. In the presence of the charged tryptophanyl-tRNATrp, transcription is attenuated due to formation of a stem-loop structure in the mRNA. Pyridoxal-phosphate is a cofactor for the tryptophan synthase enzyme.
Representation of two-component signal transduction systems
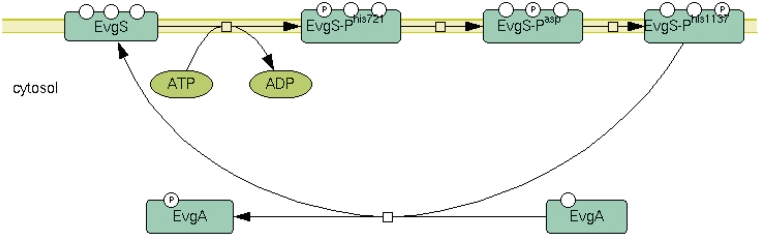
We have recently updated the representation and curation of the 26 known E. coli two-component signal transduction systems in EcoCyc. Two-component systems couple the detection of environmental signals to regulatory responses based on a phosphotransfer reaction from a histidine protein kinase to a response regulator protein (9). The previous representation of two-component systems in EcoCyc was relatively unintuitive and provided little or no information as to the sites of phosphorylation, the localization of the components, or the environment signals. In the case of complex reactions, the previous representation was also potentially misleading. Additionally, no curated summary information was previously provided for the two-component system pathways.
Figure 3 gives an example of the new representation of a signal transduction pathway in EcoCyc. EvgS is a member of the ‘unorthodox’ sensor kinase class, in which a multi-step phosphorelay occurs between alternating histidine and aspartate residues before the EvgA response regulator is activated (10). In the new depictions, each phosphorylation step and the protein localizations are much more intuitively comprehensible. The updated representations also include the environmental signal, if known, and a curated summary providing pertinent information on the system based on the published literature.
Figure 3.
New graphic representation of the EvgSA two-component signal transduction pathway.
Enhancements to the cellular and regulatory overviews
Overviews are genome-scale diagrams that can be accessed from the ‘Tools’ section in the menu bar. When an overview is displayed, a new menu containing search and highlighting options appears in the menu bar. The EcoCyc metabolic map diagram (cellular overview) has been completely reimplemented using modern Web technologies to create an interactive metabolic map that is zoomable, searchable and can be overlaid with high-throughput experimental datasets. Zooming is activated by double-clicking the area of interest, or by using the zoom ladder at the upper left. The diagram can be moved horizontally or vertically by clicking and dragging. Users can search the diagram for specific elements using the Cellular Overview menu commands, such as Highlight Gene(s) and Highlight Compound(s). Each highlight command enables entering an exact or partial name to search for, and the resulting genes or compounds are then highlighted in the diagram. Each search operation adds a new ‘overlay’ (a set of highlighted regions in the diagram). The Layer Switcher panel in the upper right presents the set of current overlays. Clicking the List button generates a list of all objects within that highlighting layer in a new window; the user can then click objects on the list to locate them in the Cellular Overview.
Highlighting patterns can be captured and saved using the command Generate Bookmark for Current Cellular Overview, which generates a URL that will recreate the highlighting. Users can also generate URLs that will highlight desired components of the diagram, including the specification of color values, as described in Cellular Overview → Help. Larger omics datasets can be painted onto the diagram interactively via a data file, or programmatically through the Web using a Web POST operation. This new implementation supports the animated display of omics data on the metabolic map, as well as zooming of the diagram with omics data present.
The regulatory overview diagram depicting the regulatory network of E. coli, which was previously available only through the desktop version of EcoCyc, is now also accessible via the Web. Its three concentric rings depict (from the inside) global E. coli regulators, other regulators and non-regulatory genes with known regulators. Currently, this diagram depicts the regulation of transcription initiation only, but this limitation will be remedied in the future. The user can right-click on a gene to show arrows to all genes that it regulates, or to all genes that regulate it. Similarly to the cellular overview, the regulatory overview is zoomable and draggable, and can be painted with omics data.
Comparative analyses
EcoCyc describes E. coli K-12. The larger BioCyc site of which EcoCyc is a part contains databases built from more than 1000 genomes, including 31 E. coli genomes and eight Shigella genomes, and more will be added on a regular basis. Click the word ‘change’ in the Quick Search area to select a different genome for querying. The same tools used to search and navigate EcoCyc, such as the genome browser, work for these other genomes. Further, several comparative tools are available to compare genome, pathway and other information (see Tools → Comparative Analysis), including a comparative genome browser that aligns chromosomal regions around orthologous genes (from a gene page, click the button Align in Multi-Genome Browser part way down the page). Note that ortholog data is not currently available for all genomes, but this limitation will be rectified in 2011. The BioCyc Guide (http://biocyc.org/BioCycUserGuide.shtml) describes how genomes are processed for inclusion in BioCyc, and how ortholog data are derived.
Web services
Much of the EcoCyc data can now be programmatically retrieved from the EcoCyc web site in XML format using web services. A diverse set of services has been made available to facilitate different types of queries. The following list briefly enumerates the currently available services. Additional services may be added in the future.
Search for a list of all gene and protein objects whose name contains the query string. For each matching gene or protein, a link to the data page for that object is provided. Examples: http://biocyc.org/ECOLI/keywordsearch?keyword=trp http://biocyc.org/ECOLI/keywordsearch?keyword=trpA&detail=T
Given a pathway ID, return the pathway data in BioPAX format (11) (either levels 2 or 3). Example: http://biocyc.org/ECOLI/pathway-biopax?type=3&object=TRPSYN-PWY
Given an object ID of any kind (such as a gene, protein, compound, reaction or pathway), return the data associated with that object in a format that corresponds closely to that of the underlying Pathway Tools schema. Where the representation of an object references other objects (such as a reaction referencing its substrate compounds), URLs are supplied that enable retrieval of those objects as well. Examples: http://biocyc.org/getxml?ECOLI:EG11025 http://biocyc.org/getxml?ECOLI:GLYCOLYSIS
Given an arbitrary query constructed using the BioVelo query language (12), return the data associated with all the matching objects. The following example returns data for all EcoCyc pathways: http://biocyc.org/xmlquery?object=[x:x<-ECOLI^^Pathways]
ENHANCEMENTS TO THE Pathway Tools DESKTOP SOFTWARE
Object groups
A new feature called Object Groups enables storing and manipulating groups of objects of interest, such as a list of genes (e.g. those up-regulated in a microarray experiment or those found in a screening experiment), a list of pathways or a list of metabolites. A scientist can define as many groups as they like, and groups can, in turn, be grouped within folders. A group can be defined by typing in a list of gene (or other object) names, by reading a list of names from a file, and from the result of an earlier query to EcoCyc. Groups are saved across executions of the software. In the future, Object Groups will also be available through the EcoCyc web site.
Groups can be displayed as a table, and a group can be highlighted on any of the three overviews. Groups can also be transformed, creating a new group derived from a previous group. For example, a gene group can be transformed to the set of pathways in which those genes occur, or to the set of all genes in all operons containing those genes, or to the set of genes that regulate the first gene set. Similarly, a pathway can be transformed to the set of all genes within the pathway, and to the set of all metabolites within the pathway.
Enrichment analysis
A group can be subject to enrichment analysis to detect overrepresented categories within the group. Analyzing for enrichment of GO terms is a commonly used method for interpreting gene expression data. For example, we can start by defining a gene group of the 45 genes from a microarray experiment involving growth of E. coli under excess tryptophan (13) whose relative expression levels are greater than a value of 2.0. We can then perform an enrichment analysis on the resulting gene group to determine whether the group is statistically overrepresented for categories of genes defined by GO. That is, does the group contain more genes within certain GO categories than would be expected by chance, and at what P-value? When applied to the tryptophan dataset, the program found that 16 of those genes were in the GO category ‘GO:0044271—cellular nitrogen compound biosynthetic process’, which was overrepresented with a P-value of 2.09E−7. The software can also search for overrepresentation of genes from EcoCyc metabolic pathways, and for overrepresentation of genes directly regulated by EcoCyc TFs. In the preceding example, the software found the set was enriched for genes in the EcoCyc superpathway for sulfate assimilation and cysteine biosynthesis with a P-value of 1.33E−5, and that the set was enriched for genes in the ArgR regulon with a P-value of 1.31E−4.
Omics data graphing
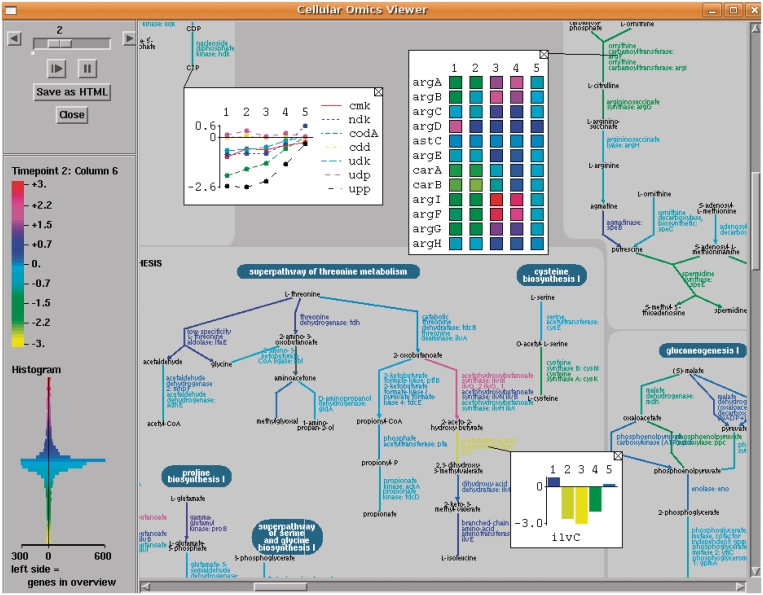
The Omics Viewers have the ability to show a graph of the set of data values for a given object in a small popup overlay superimposed on the display. For example, with a gene expression time-series experiment, the omics graph for a gene would show how the expression value for that gene changes over time. The graph for a reaction would show the changes in expression for all genes that catalyze the reaction, and the graph for a pathway would show the changes in expression for all genes that participate in the pathway. The pop-up can be customized to show the data either as a heat map, a bar graph or an x−y plot. The user can drag the popup to reposition it as desired (an optional connecting line links the popup to the object it describes). An example is shown in Figure 4. In addition, the software remembers the dataset most recently loaded into the Omics Viewers, so that these popups can then be added to any object display (such as to genes in an individual pathway display or in the genome browser display). This facility enables viewing omics data for particular objects of interest at any time, not just in the context of the Omics Viewers.
Figure 4.
Omics Data Graphing.
USING EcoCyc AS A TEACHING RESOURCE
A set of web-based undergraduate-student educational materials was developed by one member of our team (RPG) for EcoCyc. The intended student audience includes microbiology/biology/biochemistry majors, allied health majors, biotechnology majors, engineering majors and non-declared majors. The underlying goal was to explore ways to complement classroom lectures with out-of-class projects that are easily accomplished by students having little or no prior database experience. The designed activities are appropriate for the classroom, laboratory, and out-of-class activity.
Why do this? Few materials are available that support instructor-led instruction or independent student learning from the EcoCyc database. Currently, beginning microbiology students must learn by themselves to navigate the existing EcoCyc web site, a tool that is prmarily oriented towards the advanced microbiology researcher.
The student-based EcoCyc learning materials were incorporated in the UCLA ‘Introductory Microbiology 101’ course for ∼250 undergraduate students in the Spring Quarters of 2008 and 2009. Applications included research-level exercises to query and resolve a panel of genome/gene/protein/enzyme/gene regulation problems using the EcoCyc resource. Each class module contained 10–12 exercises along with a set of tutorial prompts to aid the student in researching the targeted area of E. coli biology. Following the web-based research phase, the student then generated a set of succinct replies to answer the assigned tasks. The student also provided a brief written statement explaining the rationale and approach used. The ultimate goal of this project was to stimulate inquiry-based student learning by using state-of-the-art research tools.
Using EcoCyc in an undergraduate university setting has enabled achieving far greater depth and breadth of material coverage as compared to using a lecture-only format. Student performance met or exceeded the original course expectations. Student feedback was quite positive given that this was often a first exposure to using this type of web-based organism database. Students responded that it significantly facilitated access to web content and the mastery of user tools. In short, using EcoCyc significantly enhanced the learning of underlying principles of E. coli biology.
In the future, we plan to author additional class modules and exercises in areas not currently covered. New modules will cover E. coli chemotaxis, signaling, cell division and protein export. We also plan outreach to other institutions by sharing course materials and assisting in classroom implementation.
DATABASE AVAILABILITY
The EcoCyc, MetaCyc and BioCyc databases are freely and openly available to all. See http://biocyc.org/download.shtml for download information. New versions of the downloadable data files and of the EcoCyc Web site are released four times per year.
FUNDING
National Center for Research Resources (NCRR) and National Institute of General Medical Sciences (NIGMS) (Award Number GU24GM077678-20 to P.D.K.); NIGMS (Award Number GM071962 to J.C.-V.). Funding for open access charge: National Institutes of Health (GU24GM077678).
Conflict of interest statement. SRI authors benefit from a commercial licensing program for Pathway Tools. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institute of General Medical Sciences of the National Institutes of Health.
REFERENCES
- 1.Muller HM, Kenny EE, Sternberg PW. Textpresso: an ontology-based information retrieval and extraction system for biological literature. PLoS Biol. 2004;2:e309. doi: 10.1371/journal.pbio.0020309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Gama-Castro S, Jimenez-Jacinto V, Peralta-Gil M, Santos-Zavaleta A, Penaloza-Spinola MI, Contreras-Moreira B, Segura-Salazar J, Muniz-Rascado L, Martinez-Flores I, Salgado H, et al. RegulonDB (version 6.0): gene regulation model of Escherichia coli K-12 beyond transcription, active (experimental) annotated promoters and Textpresso navigation. Nucleic Acids Res. 2008;36:D120–D124. doi: 10.1093/nar/gkm994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Thomas-Chollier M, Sand O, Turatsinze JV, Janky R, Defrance M, Vervisch E, Brohee S, van Helden J. RSAT: regulatory sequence analysis tools. Nucleic Acids Res. 2008;36:W119–W127. doi: 10.1093/nar/gkn304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pedersen H, Valentin-Hansen P. Protein-induced fit: the CRP activator protein changes sequence-specific DNA recognition by the CytR repressor, a highly flexible LacI member. EMBO J. 1997;16:2108–2118. doi: 10.1093/emboj/16.8.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jorgensen CI, Kallipolitis BH, Valentin-Hansen P. DNA-binding characteristics of the Escherichia coli CytR regulator: a relaxed spacing requirement between operator half-sites is provided by a flexible, unstructured interdomain linker. Mol. Microbiol. 1998;27:41–50. doi: 10.1046/j.1365-2958.1998.00655.x. [DOI] [PubMed] [Google Scholar]
- 6.The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hu JC, Karp PD, Keseler IM, Krummenacker M, Siegele DA. What we can learn about Escherichia coli through application of Gene Ontology. Trends Microbiol. 2009;17:269–278. doi: 10.1016/j.tim.2009.04.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2010;38:D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gao R, Stock AM. Biological insights from structures of two-component proteins. Annu. Rev. Microbiol. 2009;63:133–154. doi: 10.1146/annurev.micro.091208.073214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Appleby JL, Parkinson JS, Bourret RB. Signal transduction via the multi-step phosphorelay: not necessarily a road less traveled. Cell. 1996;86:845–848. doi: 10.1016/s0092-8674(00)80158-0. [DOI] [PubMed] [Google Scholar]
- 11.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nat. Biotech. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Latendresse M, Karp PD. An advanced web query interface for biological databases. Database. 2010 doi: 10.1093/database/baq006. 2010, doi:10.1093/database/baq006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Khodursky AB, Peter BJ, Cozzarelli NR, Botstein D, Brown PO, Yanofsky C. DNA microarray analysis of gene expression in response to physiological and genetic changes that affect tryptophan metabolism in Escherichia coli. Proc. Natl Acad. Sci. USA. 2000;97:12170–12175. doi: 10.1073/pnas.220414297. [DOI] [PMC free article] [PubMed] [Google Scholar]