Abstract
Now in its 10th year, the Gramene database (http://www.gramene.org) has grown from its primary focus on rice, the first fully-sequenced grass genome, to become a resource for major model and crop plants including Arabidopsis, Brachypodium, maize, sorghum, poplar and grape in addition to several species of rice. Gramene began with the addition of an Ensembl genome browser and has expanded in the last decade to become a robust resource for plant genomics hosting a wide array of data sets including quantitative trait loci (QTL), metabolic pathways, genetic diversity, genes, proteins, germplasm, literature, ontologies and a fully-structured markers and sequences database integrated with genome browsers and maps from various published studies (genetic, physical, bin, etc.). In addition, Gramene now hosts a variety of web services including a Distributed Annotation Server (DAS), BLAST and a public MySQL database. Twice a year, Gramene releases a major build of the database and makes interim releases to correct errors or to make important updates to software and/or data.
INTRODUCTION
Scientific advances in genomics promise to help plant breeders improve quality, pathogen resistance, and yield to meet the growing demands for food, fiber and biofuel, however, the ever-increasing volume of sequence data generated from reference genomes, expression studies and genome-wide genetic diversity studies present challenges to efficiently store, curate, analyze and retrieve such data. Gramene is a free online database for comparative plant genomics that began as an extension of the RiceGenes project (1,2) and now holds many large and varied data sets that are used extensively by thousands of plant researchers in the public and private sectors throughout the US, Asia and Europe. Through the application of standardized annotation methods, Gramene strives to create a resource that promotes cross-species analysis of both conserved and species-specific functions. Various ontologies are used to consistently describe plant anatomy (3), phenotype traits (4), genes (5), environment and taxonomy, and both computational and manual curation are employed to integrate data sets from various leading research projects on plants and public repositories such as GenBank. This article summarizes the changes to the website since the last publication in NAR 2008 (6), through the 31st release of the Gramene website in May 2010.
GENOMES
Plant biologists often enter Gramene through their species of interest, and genome browsers offer a direct window on specific regions and genes. Since Gramene’s inception, we have used the Ensembl genome browser (7). As of an interim release made shortly after our May 2010 release, Gramene uses Ensembl version 58 to visualize eight complete and several more partial plant genomes available from http://www.gramene.org/genome_browser/. Annotations held by Gramene include ab initio, evidence-based and community-generated gene predictions, repeat regions, and homology as well as cross-references to sequences in public databases, locations of quantitative trait loci (QTLs), locations of microarray probes, cross-references to sequences in public databases and genome variation such as SNPs and indels. The generation of genome annotations has been described previously (8). Each release of the database contains new and updated annotations. Since our last publication, Gramene has added or updated many plant genomes listed in Table 1.
Table 1.
A listing of the whole genomes available in Gramene
| Oryza sativa japonica | Updated to MSU version 6 released in January 2009 (33) with 160 000 SNPs from 20 O. sativa lines determined as part of the OryzaSNP project using SNP array technology (34) |
| Oryza sativa indica | The Beijing Genome Institute (BGI) assembly of cultivar 93-11 published in 2005 (35) |
| Arabidopsis thaliana | Updated to The Arabidopsis Information Resource (TAIR) (36) version 9 released in June 2009 with the Ensembl database created by the Nottingham Arabidopsis Stock Centre (NASC) |
| 637 522 SNPs from 20 A. thaliana lines determined as part of the Arabidopsis 2010 project using genome tiling array technology | |
| 220 000 SNPs from 363 A. thaliana lines determined as part of the Arabidopsis 2010 project using SNP array technology | |
| 2 698 797 SNPs from 17 A. thaliana lines determined as part of the Arabidopsis 1001 genomes project using re-sequencing technology | |
| Arabidopsis lyrata | Added the Araly1 assembly from the Joint Genomes Institute (JGI) |
| Brachypodium distachyon | Added the Brachy 1.2 version from JGI (2010) |
| Populus trichocarpa | Added JGI version 2.0 assembly (January 2010) and JGI version 2.0 gene predictions (March 2010) (37) |
| Sorghum bicolor | Added the Sbi1 assembly and Sbi1.4 gene set (March 2007) (38) |
| Vitis vinifera | Added the International Grape Genome Program (IGGP) and version ‘IGGP 12X’ (39) with 469 470 SNPs from 17 V. vinifera lines determined as part of the USDA project using re-sequencing technology (40) |
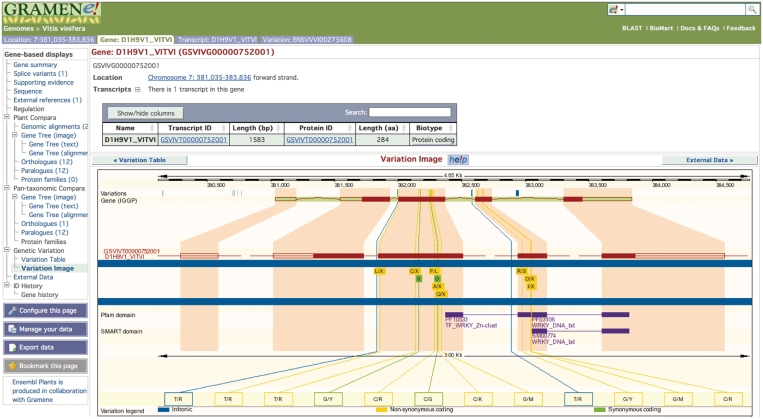
In addition to the fully sequenced genomes, Gramene has worked with the Oryza Mapping Alignment Project (OMAP) (9) to visualize the physical map of O. rufipogon and the chromosome 3 short arms of O. brachyantha, O. nivara, O. rufipogon, O. barthii, O. glaberrima, O. minuta CC, O. officinalis and O. punctata. We have also now integrated variation data into our genomes such as a set of 71K single nucleotide polymorphisms (SNPs) from grape (10) in order to help researchers to determine the consequence of variation (Figure 1). The Arabidopsis variation database contains data from the screening of over 900 strains using the Affymetrix 250k Arabidopsis SNP chip (http://walnut.usc.edu/2010/data/250k-data-version-3.04) as well as SNP discovery data used to construct the 250K chip from 20 re-sequenced Arabidopsis lines (11).
Figure 1.
An Ensembl browser view showing Vitis vinifera SNPs in the context of gene annotation. SNPs are color-coded to indicate position relative to gene features (e.g. `intronic') and consequences of SNP on coding sequence (e.g. `non-synonymous').
In 2009, Gramene entered into a formal collaboration with the European Molecular Biology Laboratory - European Bioinformatics Institute (EMBL-EBI) and their Ensembl Genomes (EG) project (12) to create a common set of databases and annotations. Gramene has contributed all the ‘core’ databases for the fully sequenced plant genomes available at EG website (http://plants.ensembl.org), and both groups work on quality control, the integration of content, and the development of new features to share across all available plant genomes, thereby reducing redundancy of effort and standardizing analyses and visualization for the community.
WHOLE GENOME ALIGNMENTS
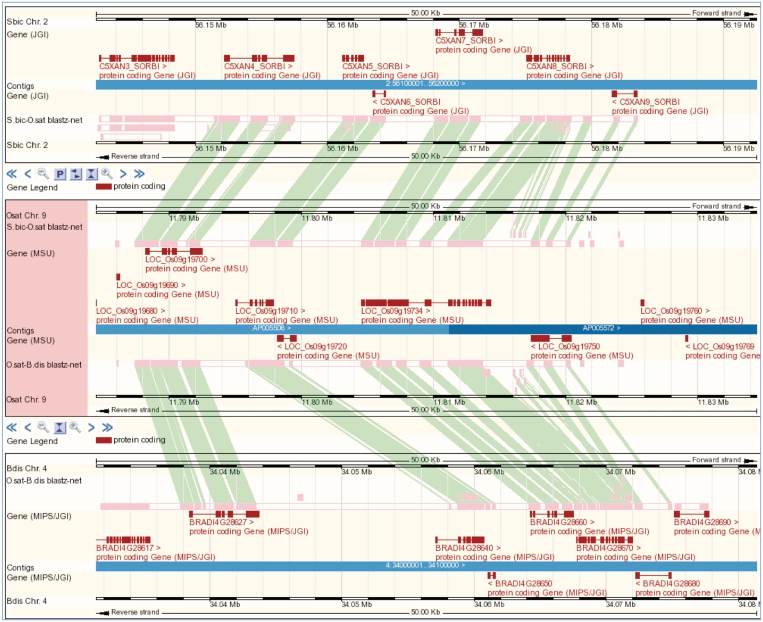
Researchers are often use whole genome alignments (WGA) to explore conservation of chromosomal structure and gene structure. Gramene provides pre-computed whole genome and gene–gene alignments using a BLASTZ-net pairwise (13,14) whole genome alignment method implemented by Ensembl to analyze 12 plant genomes (http://www.gramene.org/info/docs/compara/analyses.html#blastz). Ensembl’s release 56 reintroduced multi-species comparative genome views driven by pair-wise alignments that had been absent from the Ensembl views for a year. Figure 2 gives an example showing homology from a 50 Kb region on O. sativa japonica chromosome 9 (central panel) showing and similar sized regions of Sorghum bicolor chromosome 2 (top panel) and Brachypodium distachyon chromosome 4 (bottom panel).
Figure 2.
The new multi-species view shows alignments in the context of gene annotations across multiple species. In this case, a region of rice (center) is displayed against homologous regions in sorghum (top) and Brachypodium (bottom). To create such a view from any location-based display, a user would select the `multi-species view' option from the navigation hierarchy. Referent species can be added to and removed from the display using the `select species' option.
GENE TREES
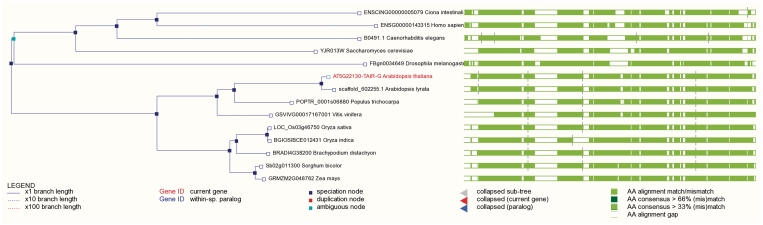
Comparative functional genomics allows researchers to trace evolutionary histories of genes and traits, and Gramene's Compara database adds a new level of tools to help researchers make inferences of function and strategies for gene annotation. Gramene uses the standard Ensembl GeneTree method (15) to generate gene trees and predict ortholog and paralog relationships between species. In the current release, the GeneTree database was rebuilt using five monocot genomes (O. sativa japonica, O. sativa indica, O. glaberrima, B. distachyon and S. bicolor), four dicot genomes (A. lyrata, A. thaliana, P. trichocarpa and V. vinifera) and five model metazoan genomes (Caenorhabditis elegans, Ciona intestinalis, Drosophila melanogaster, Homo sapiens and Saccharomyces cerevisiae). Figure 3 shows an example of the results of our latest gene tree build.
Figure 3.
Phylogenetic tree for Arabidopsis gene PNT1, a Glycosyltransferase, showing conservation throughout the eukaryotic lineage.
COMPARA AND SYNTENY ANALYSIS
Synteny analysis allows researchers to infer ancestral locations of genes, and the finding of conserved synteny provides a measure of confidence that genes are true orthologs. In previous builds, Gramene used DNA-level whole genome alignments across its many hosted genomes, but, in the current release, Gramene implemented a new synteny analysis pipeline that makes use of gene ortholog assignments from our Compara GeneTree output as additional parameter to confirm homology. This avoids the complications associated with using WGA including spurious alignment and differential expansion and contraction within and between genomes. The new method was originally developed for the Maize Project (16) and is now implemented as a ‘runnable’ within our standardize genome annotation methods (17). To start the analysis, strictly collinear orthologs are mapped using DAGchainer (18) giving rise to the classification of high-confidence ‘syntenic:collinear’ gene-pairs. Next these mappings are used as anchor points to identify additional syntenic orthologs that may violate collinearity due to local rearrangements or assembly artifacts. This step is configured using a gene-index distance parameter, and its output defines near-collinear gene pairs classified as ‘syntenic:in-range’. These relationships are stored as gene attributes, and ranges of syntenic blocks are displayed with the Ensembl SyntenyView module. Table 2 shows the three pairs of genomes compared in release 31.
Table 2.
Pair-wise synteny analysis available in Gramene
| Oryza sativa japonica | Oryza sativa japonica | ||
| Sorghum bicolor | Yes | Sorghum bicolor | |
| Brachypodium distachyon | Yes | Yes | Brachypodium distachyon |
PATHWAYS
Gramene hosts metabolic pathway databases for eight species including rice, sorghum, Arabidopsis (19), tomato, potato, pepper, Medicago (20), coffee, as well as three reference databases, EcoCyc (21), PlantCyc (22) and MetaCyc (23). These display gene functions in the context of biochemical reactions and networks. Users can download lists of genes associated with each pathway and extract inter-specific comparisons between pathways and associated genes. Gene identifiers link to the gene summary pages of Gramene’s Ensembl genome browser, and we have added an ‘Omics Validator’ tool to map user-provided microarray probe identifiers from various microarray platforms to their respective gene identifiers, starting with rice. The mappings for the arrays are provided from the functional genomics module in the genome browser.
In the current release of the rice pathway database developed by Gramene, our curators added approximately 170 enzymatic and 80 transport reactions, revised approximately 65 tRNA and 600 transport reaction-associated genes, and updated several important rice pathways. Gramene’s RiceCyc has 342 known or predicted metabolic pathways for O. sativa japonica cultivar ‘Nipponbare’ and has undergone several rounds of data-quality enhancement and manual curation. More than 100 literature citations were added or curated. The first release of the Sorghum metabolic pathways (SorghumCyc) developed by Gramene provides 328 pathways. The pathways from rice and sorghum, both developed by Gramene, are provided in a web-based browsable form as well as for bulk download in several options including the BioPax (24) and Systems Biology Markup Language (SBML) (25) formats for advanced users. The annotated pathways are used as external references in the sorghum and rice genome browsers.
GENETIC DIVERSITY
Manipulating and storing vast amounts of sequence data from increasingly cheaper and faster sequencing methodologies is a significant challenge. Gramene’s genetic diversity module is specifically designed to facilitate the integration and analysis of these data. It uses the Genomic Diversity and Phenotype Data Model (GDPDM, http://www.maizegenetics.net/gdpdm/) to store RFLP, SSR and SNP allele data, information about QTL, and passport data for wild and cultivated germplasm from rice, maize, wheat, Arabidopsis, and sorghum along with quantitative phenotypic data for some genotype accessions (Table 3).
Table 3.
Large-scale variation-based genotype data sets available in Gramene’s genetic diversity database
| Rice | OryzaSNP large scale SNP variation study (41) (∼160 K SNPs × 20 diversity rice accessions), mapped from IRGSP4 to MSU6 |
| Maize | Panzea SNP data (1.6MSNPs × 27 NAM founder lines) |
| Arabidopsis | 2010 Project SNP discovery (42) (637 522 SNPs, 20 accessions), mapped from TAIR8 to TAIR9 |
| 2010 Project genotype data v3.04 (∼214K SNPs × 1179 Arabidopsis accessions), mapped from TAIR8 to TAIR9. Construction of 250K chip used in this study is discussed in Clark (42) and Kim (43) | |
| 1001 Genomes WTCHG/Mott data from dbSNP (2 698 797 SNPs, 17 accessions) |
In 2010, the GDPDM schema was updated to include a data packing system that can easily store and quickly retrieve millions of SNPs. By using binary large objects (BLOBs) in the database, we reduced the space required to store variation data by several orders of magnitude, thereby allowing us to easily query many large data sets. Gramene’s new SNP Query tool (Figure 4) uses this improvement to quickly retrieve and filter SNP data by chromosome and cultivar subgroups. The results provide information about overlapping genomic features and links to visualize them in the Ensembl genome browser. We now provide data sets for visualizing genotype patterns across cultivars of interest using the Scottish Crop Research Institute’s Flapjack program (http://bioinf.scri.ac.uk/flapjack/). A Java Web Start-enabled version of the Tassel (26) program is provided for evaluating trait associations, patterns of linkage disequilibrium and genetic diversity. In the last year, we have added many features to Tassel including a new alignment viewer, progress monitoring, pipelines and wizards for automatic data loading and analysis. For users who prefer to interact with data using their own tools, all diversity data is provided in various download formats including HapMap and PLINK at http://www.gramene.org/diversity/download_data.html.
Figure 4.
The new SNP Query tool returns variation from one or more accessions based on genomic coordinates. The `genes' column contains hyperlinks to the Ensembl genome browser’s gene summary page.
GERMPLASM
A new entry point for plant breeders and geneticists was added by way of the ‘germplasm’ unit (http://www.gramene.org/db/germplasm/) to summarize all the curated data we hold for the most popular cultivars and wild accessions of rice. Access to this database is by species or genotype/germplasm accession instead of genomic coordinates or markers. From the germplasm home page, users can search for markers or genetic diversity information related to a particular accession.
MARKERS, SEQUENCES AND MAPS
In addition to the many custom data sets we curate in collaboration with researchers in the plant community, Gramene mirrors GenBank’s Viridiplantae sequences for our genome alignment pipeline. Gramene’s markers and sequences database now holds around 49-million records we judge to be the most valuable to our users. This database also stores the results of the alignments from our annotation results for our completed genomes as well as manually curated maps provided by the researchers/projects and those extracted from peer-reviewed publications. As this database is also the source for Gramene’s comparative maps and DAS, it is a central organizing point for users to see how markers and sequences are related to each other as well as to QTLs, source germplasm and various ontologies.
Gramene’s comparative maps database now holds almost 8M features on 214 map sets from genetic, physical, bin, sequence, cytogenetic and QTL studies. Gramene uses the CMap application (27) to allow users to create cross-species comparisons of any map type. Since last publication, we have curated from literature an additional 17 maps from rice, sorghum, barley, maize, wheat and Aegilops tauschii (28) as shown in Figure 5. Links from CMap’s feature details page allow the user to return to the source markers and sequences database to explore associations to other data sets in Gramene such as ontologies and genes.
Figure 5.
A comparative map view showing the genetic map of Aegilops tauschii along side the latest O. sativa japonica sequence map and a rice QTL map.
QTL
Gramene’s QTL database (29) has seen no change to the number of QTL since our last update, holding steady at 11 624 curated QTL from 10 species. The QTL are associated to terms from trait ontology (TO), plant ontology (PO), growth ontology (GRO), environment ontology (EO), as well as to co-localized or neighboring markers and Gramene gene identifiers. A recent improvement is that users may now search for QTL by any of these associations. By following links to the various ontology term definitions, users may see genes, proteins, markers and other QTL also related to the term. The locations of rice QTL on the O. sativa japonica genome are inferred through the alignments of their associated markers. Links from the QTL details pages allow the user to view QTL on the experimental map in CMap or in the Ensembl browser where the ‘Export data’ button allows users to easily extract all the features (genes, repeats, SNPs, etc.) located in the QTL’s region.
INFRASTRUCTURE, QUICK SEARCH AND GRAMENE MART
Since our last update, we have continued to work on making our user interfaces cleaner and more informative. Our footer bar was redesigned to be smaller and less obtrusive, and the front page was redesigned to highlight Gramene’s major data sets (e.g. genes, proteins, QTL) as entry points for users (Figure 6). Also prominently featured on the front page as well as in the upper-right corner of every page is the ‘quick search’ which has itself been improved with the ability to filter results by species where applicable. For bioinformatics and software developers interested in installing a local copy of the Gramene database, we upgraded the internal web server to the most recent Apache version 2. Gramene also hosts several BioMart databases to allow users to easily execute complex queries of various data sets we hold, the results of which can be viewed in the web browser, downloaded, or integrated into the Galaxy system (30).
Figure 6.
Gramene’s redesigned home page allows quick access to all our major data sets and quick search.
WEB SERVICES
Sometimes the advanced user needs access to Gramene’s data through means other than our web pages, so we provide several ways to directly connect such as our public, read-only MySQL server. The host ‘gramenedb.gramene.org’ mirrors the current build of our databases and can be accessed using the password ‘gramene.’ With over 300 tracks to choose from, Gramene’s DAS can be used with our Ensembl browsers or any other DAS client to access our annotations. Recently we improved the query engine by moving from MySQL to FastBit (31), a bitmap indexing system that executes queries in a fraction of the time from MySQL. The aforementioned GDPC API also allows direct interaction with our diversity databases. Finally, Gramene continues to maintain BLAST databases for our users.
THIRD-PARTY SUBMISSION OF DATA
In an effort to encourage community curation, Gramene created the PlantGeneWiki (http://plantgenewiki.gramene.org/) to allow users to search genes as well as to register and contribute new and edit existing genes from plant species. Designed as an online community portal on plant genes and their annotations, the site is managed by the research community and Gramene staff.
DATA AND SOFTWARE AVAILABILITY
Gramene makes all databases and software freely available under the GNU General Public License. Downloads are available from the Gramene FTP site (ftp://ftp.gramene.org). In addition, Gramene allows anonymous, read-only access to the Subversion source code repository at http://svn.warelab.org/gramene/trunk. In this way, users can have access to any previous release as well as the most current changes in our development code.
OUTREACH
The Gramene staff uses many methods to inform, educate and interact with our users. A public news blog (http://news.gramene.org) with RSS feed capabilities is maintained to keep our users informed of changes to the website as well as important publications, job opportunities and meetings of interest to our researchers. In addition to our on-going relationship with OpenHelix (http://www.openhelix.com) (32) to provide tutorials, in the last year members of the Gramene team have been creating very short video tutorials that introduce very specific topics on Gramene or new tools and data sets (http://www.gramene.org/tutorials). Our staff also presents posters, talks and hands-on workshops at meetings such as the annual Plant and Animal Genome (PAG) conference, the Rice Technical Working Group, the Maize Genetics Meeting, Intelligent Systems for Molecular Biology (ISMB), Plant Biology and Genome Informatics.
FUNDING
National Science Foundation (0703908, 0851652). Funding for open access charge: National Science Foundation (0321685); NSF DBI (0703908).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank our users for their feedback and support as well as our collaborators and contributors who have supplied Gramene with data, especially NSF projects #0638566 (High Density Scoreable Markers for Maize Trait Dissection), #0321538 (An Annotation Resource for the Rice Genome), #0606461 (Exploring the Genetic Basis of Transgressive Variation in Rice), #0723510 (Collaborative Research: An Arabidopsis Polymorphism Database), #0723510 (Collaborative Research: An Arabidopsis Polymorphism Database), #0638820 (OMAP), #0701916 (Physical Mapping of the Wheat D Genome), #0743804 (POPcorn), #0543441 (NextGen PLEXdb), #0638820 (The evolutionary genomics of invasive weedy rice) and the USDA-ARS CRIS 9235-21000-013-00D (Complete Switchgrass Genetic Maps Reveal Subgenome Collinearity, Preferential Pairing and Multilocus). Gramene is deeply indebted to our Science Advisory Board members Paul Flicek, Michael Ashburner, Anna McClung, Georgia Davis, David Marshall, Patricia Klein, William Beavis, Tim Nelson for their critical comments, suggestions and improvements. We also thank Peter Van Buren for his excellent system administration work.
REFERENCES
- 1.McCouch SR, Paul E. RiceGenes, an International Genome Database and Bulletin Board for Rice. DNA Link. 1993;3:40–41. [Google Scholar]
- 2.Ware D, Jaiswal P, Ni J, Pan X, Chang K, Clark K, Teytelman L, Schmidt S, Zhao W, Cartinhour S, et al. Gramene: a resource for comparative grass genomics. Nucleic Acids Res. 2002;30:103–105. doi: 10.1093/nar/30.1.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Jaiswal P, Avraham S, Ilic K, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, Schaeffer M, et al. Plant Ontology (PO): a controlled vocabulary of plant structures and growth stages. Comp. Funct. Genomics. 2005;6:388–397. doi: 10.1002/cfg.496. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Yamazaki Y, Jaiswal P. Biological ontologies in rice databases. An introduction to the activities in Gramene and Oryzabase. Plant Cell Physiol. 2005;46:63–68. doi: 10.1093/pcp/pci505. [DOI] [PubMed] [Google Scholar]
- 5.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, Hurwitz B, McCouch S, Ni J, Pujar A, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36(Database issue):D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl’s 10th year. Nucleic Acids Res. 2009;38(Database issue):D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang C, Jaiswal P, Hebbard C, Avraham S, Buckler ES, Casstevens T, Hurwitz B, McCouch S, Ni J, Pujar A, et al. Gramene: a growing plant comparative genomics resource. Nucleic Acids Res. 2008;36(Database issue):D947–D953. doi: 10.1093/nar/gkm968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wing RA, Ammiraju JS, Luo M, Kim H, Yu Y, Kudrna D, Goicoechea JL, Wang W, Nelson W, Rao K, et al. The oryza map alignment project: the golden path to unlocking the genetic potential of wild rice species. Plant Mol. Biol. 2005;59:53–62. doi: 10.1007/s11103-004-6237-x. [DOI] [PubMed] [Google Scholar]
- 10.Myles S, Chia JM, Hurwitz B, Simon C, Zhong GY, Buckler E, Ware D. Rapid genomic characterization of the genus vitis. PLoS One. 2010;5:e8219. doi: 10.1371/journal.pone.0008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 12.Kersey PJ, Lawson D, Birney E, Derwent PS, Haimel M, Herrero J, Keenan S, Kerhornou A, Koscielny G, Kahari A, et al. Ensembl Genomes: extending Ensembl across the taxonomic space. Nucleic Acids Res. 2010;38(Database issue):D563–D569. doi: 10.1093/nar/gkp871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Schwartz S, Kent WJ, Smit A, Zhang Z, Baertsch R, Hardison RC, Haussler D, Miller W. Human-mouse alignments with BLASTZ. Genome Res. 2003;13:103–107. doi: 10.1101/gr.809403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kent WJ, Baertsch R, Hinrichs A, Miller W, Haussler D. Evolution's cauldron: duplication, deletion, and rearrangement in the mouse and human genomes. Proc. Natl Acad. Sci. USA. 2003;100:11484–11489. doi: 10.1073/pnas.1932072100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Vilella AJ, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: Complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schnable PS, Ware D, Fulton RS, Stein JC, Wei F, Pasternak S, Liang C, Zhang J, Fulton L, Graves TA, et al. The B73 maize genome: complexity, diversity, and dynamics. Science. 2009;326:1112–1125. doi: 10.1126/science.1178534. [DOI] [PubMed] [Google Scholar]
- 17.Potter SC, Clarke L, Curwen V, Keenan S, Mongin E, Searle SM, Stabenau A, Storey R, Clamp M. The Ensembl analysis pipeline. Genome Res. 2004;14:934–941. doi: 10.1101/gr.1859804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Haas BJ, Delcher AL, Wortman JR, Salzberg SL. DAGchainer: a tool for mining segmental genome duplications and synteny. Bioinformatics. 2004;20:3643–3646. doi: 10.1093/bioinformatics/bth397. [DOI] [PubMed] [Google Scholar]
- 19.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36(Database issue):D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Urbanczyk-Wochniak E, Sumner LW. MedicCyc: a biochemical pathway database for Medicago truncatula. Bioinformatics. 2007;23:1418–1423. doi: 10.1093/bioinformatics/btm040. [DOI] [PubMed] [Google Scholar]
- 21.Keseler IM, Bonavides-Martinez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37(Database issue):D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhang P, Dreher K, Karthikeyan A, Chi A, Pujar A, Caspi R, Karp P, Kirkup V, Latendresse M, Lee C, et al. Creation of a genome-wide metabolic pathway database for Populus trichocarpa using a new approach for reconstruction and curation of metabolic pathways for plants. Plant Physiol. 2010;153:1479–91. doi: 10.1104/pp.110.157396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Caspi R, Altman T, Dale JM, Dreher K, Fulcher CA, Gilham F, Kaipa P, Karthikeyan AS, Kothari A, Krummenacker M, et al. The MetaCyc database of metabolic pathways and enzymes and the BioCyc collection of pathway/genome databases. Nucleic Acids Res. 2009;38(Database issue):D473–D479. doi: 10.1093/nar/gkp875. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Demir E, Cary MP, Paley S, Fukuda K, Lemer C, Vastrik I, Wu G, D'Eustachio P, Schaefer C, Luciano J, et al. The BioPAX community standard for pathway data sharing. Nat. Biotechnol. 2010;28:935–942. doi: 10.1038/nbt.1666. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gauges R, Rost U, Sahle S, Wegner K. A model diagram layout extension for SBML. Bioinformatics. 2006;22:1879–1885. doi: 10.1093/bioinformatics/btl195. [DOI] [PubMed] [Google Scholar]
- 26.Bradbury PJ, Zhang Z, Kroon DE, Casstevens TM, Ramdoss Y, Buckler ES. TASSEL: software for association mapping of complex traits in diverse samples. Bioinformatics. 2007;23:2633–2635. doi: 10.1093/bioinformatics/btm308. [DOI] [PubMed] [Google Scholar]
- 27.Youens-Clark K, Faga B, Yap IV, Stein L, Ware D. CMap 1.01: a comparative mapping application for the Internet. Bioinformatics. 2009;25:3040–3042. doi: 10.1093/bioinformatics/btp458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Luo MC, Deal KR, Akhunov ED, Akhunova AR, Anderson OD, Anderson JA, Blake N, Clegg MT, Coleman-Derr D, Conley EJ, et al. Genome comparisons reveal a dominant mechanism of chromosome number reduction in grasses and accelerated genome evolution in Triticeae. Proc. Natl Acad. Sci. USA. 2009;106:15780–15785. doi: 10.1073/pnas.0908195106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ni J, Pujar A, Youens-Clark K, Yap I, Jaiswal P, Tecle I, Tung CW, Ren L, Spooner W, Wei X, et al. Gramene QTL database: development, content and applications. Database. 2009 doi: 10.1093/database/bap005. doi:10.1093/bap005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Blankenberg D, Taylor J, Schenck I, He J, Zhang Y, Ghent M, Veeraraghavan N, Albert I, Miller W, Makova KD, et al. A framework for collaborative analysis of ENCODE data: making large-scale analyses biologist-friendly. Genome Res. 2007;17:960–964. doi: 10.1101/gr.5578007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wu K. FastBit: interactively searching massive data. J. Phys.: Conf. Ser. 2009;180 [Google Scholar]
- 32.Williams JM, Mangan ME, Perreault-Micale C, Lathe S, Sirohi N, Lathe WC. OpenHelix: bioinformatics education outside of a different box. Brief Bioinform. 2010 doi: 10.1093/bib/bbq026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Ouyang S, Zhu W, Hamilton J, Lin H, Campbell M, Childs K, Thibaud-Nissen F, Malek RL, Lee Y, Zheng L, et al. The TIGR Rice Genome Annotation Resource: improvements and new features. Nucleic Acids Res. 2007;35(Database issue):D883–D887. doi: 10.1093/nar/gkl976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl Acad. Sci. USA. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhao W, Wang J, He X, Huang X, Jiao Y, Dai M, Wei S, Fu J, Chen Y, Ren X, et al. BGI-RIS: an integrated information resource and comparative analysis workbench for rice genomics. Nucleic Acids Res. 2004;32(Database issue):D377–D382. doi: 10.1093/nar/gkh085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Swarbreck D, Wilks C, Lamesch P, Berardini TZ, Garcia-Hernandez M, Foerster H, Li D, Meyer T, Muller R, Ploetz L, et al. The Arabidopsis Information Resource (TAIR): gene structure and function annotation. Nucleic Acids Res. 2008;36(Database issue):D1009–D1014. doi: 10.1093/nar/gkm965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Tuskan GA, Difazio S, Jansson S, Bohlmann J, Grigoriev I, Hellsten U, Putnam N, Ralph S, Rombauts S, Salamov A, et al. The genome of black cottonwood, Populus trichocarpa (Torr. & Gray). Science. 2006;313:1596–1604. doi: 10.1126/science.1128691. [DOI] [PubMed] [Google Scholar]
- 38.Paterson AH, Bowers JE, Bruggmann R, Dubchak I, Grimwood J, Gundlach H, Haberer G, Hellsten U, Mitros T, Poliakov A, et al. The Sorghum bicolor genome and the diversification of grasses. Nature. 2009;457:551–556. doi: 10.1038/nature07723. [DOI] [PubMed] [Google Scholar]
- 39.Jaillon O, Aury JM, Noel B, Policriti A, Clepet C, Casagrande A, Choisne N, Aubourg S, Vitulo N, Jubin C, et al. The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature. 2007;449:463–467. doi: 10.1038/nature06148. [DOI] [PubMed] [Google Scholar]
- 40.Myles S, Chia JM, Hurwitz B, Simon C, Zhong GY, Buckler E, Ware D. Rapid genomic characterization of the genus vitis. PLoS One. 5:e8219. doi: 10.1371/journal.pone.0008219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl Acad. Sci. USA. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 43.Kim S, Plagnol V, Hu TT, Toomajian C, Clark RM, Ossowski S, Ecker JR, Weigel D, Nordborg M. Recombination and linkage disequilibrium in Arabidopsis thaliana. Nat. Genet. 2007;39:1151–1155. doi: 10.1038/ng2115. [DOI] [PubMed] [Google Scholar]