Abstract
Under the International Nucleotide Sequence Database Collaboration (INSDC; http://www.insdc.org), globally comprehensive public domain nucleotide sequence is captured, preserved and presented. The partners of this long-standing collaboration work closely together to provide data formats and conventions that enable consistent data submission to their databases and support regular data exchange around the globe. Clearly defined policy and governance in relation to free access to data and relationships with journal publishers have positioned INSDC databases as a key provider of the scientific record and a core foundation for the global bioinformatics data infrastructure. While growth in sequence data volumes comes no longer as a surprise to INSDC partners, the uptake of next-generation sequencing technology by mainstream science that we have witnessed in recent years brings a step-change to growth, necessarily making a clear mark on INSDC strategy. In this article, we introduce the INSDC, outline data growth patterns and comment on the challenges of increased growth.
INTRODUCTION
The International Nucleotide Sequence Database Collaboration (INSDC; http://www.insdc.org) represents one of the most celebrated global initiatives in public domain data sharing. Growing from efforts in the early 1980s to capture and present the increasing volumes of sequence and annotation that arose from the emerging application of sequencing techniques, by 1987, the INSDC had taken shape with the stable three party membership that persists to this day. The parties to the collaboration are the DNA Databank of Japan (DDBJ) at the National Institute for Genetics in Mishima, Japan; the European Molecular Biology Laboratory’s European Bioinformatics Institute (EMBL-EBI) in Hinxton, UK; and the National Center for Biotechnology Information (NCBI) in Bethesda, Maryland, USA. Together, the INSDC partners set out to provide a globally comprehensive collection of public domain nucleotide sequence and associated metadata. Coverage includes the spectrum of data, ranging from raw reads, through assembly and alignment information, to submitted functional annotation of assembled sequences. Raw data archives under the collaboration are known as the Trace Archive for raw data from capillary electrophoresis platforms and the Sequence Read Archive [SRA, (1)] for raw and read alignment data from next-generation platforms. Assembled sequences and annotations are available from DDBJ (2), the EMBL-Bank component of the European Nucleotide Archive (3) and GenBank from NCBI (4). Routine data exchange, standard formats and, increasingly, the sharing of technology, provide global synchrony across the collaboration.
COLLABORATIVE INSTRUMENTS
The INSDC supports data exchange pipelines through the development and maintenance of a number of core collaborative instruments. The oldest of these is the INSDC Feature Table Document, in which functional annotation conventions are described at both syntactic and semantic levels. Typically updated twice a year, the most recent version is available at: http://www.insdc.org/documents/feature_table.html. Over time, this specification has defined a bioinformatics standard used well beyond the INSDC both at the level of a format for data presentation, exchange and input for analysis tools and as a starting point for the development of annotation systems and technologies based on the feature key and qualifier definitions.
A second key collaborative instrument is the unified accessioning system. Through the sharing of the accession namespace, INSDC accessions are universal across the collaborators’ services such that a single accession will return the same sequence regardless of the query site.
A third core collaborative instrument is the data model that underlies the SRA (1). In the SRA, metadata, with information relating to a sample, experimental design, library creation and machine configuration, are expressed and exchanged in a series of XML documents. The sequence read, quality and read alignment data are stored in binary data files and linked to the SRA metadata layer.
A further collaborative instrument of importance is the INSDC status convention (http://www.insdc.org/insdc_status.html), in which a consistent level of availability for given records is maintained across the INSDC partners. This system supports such concepts as fully public data, data held confidential prior to publication and data suppressed as updated improved data become available.
Finally, during 2010, significant effort has been invested in a further collaborative instrument, the developing BioProjects database, in which data providers and INSDC database curators collate top-level information that relates otherwise dispersed sequence records to coherent studies that target complete genomes, transcriptomes, metagenomics projects, targeted locus studies and many more. While INSDC partners have collected information under this initiatiave, a major new schema expected in 2011 will support data access and mining tools.
POLICY
INSDC partners operate coordinated and integrated services closely. For the data submitter, it is only necessary to provide sequence data to one of the partners. Sequences are accessioned across a single namespace such that an accession search yields the same data content regardless of which partner institute has provided the search facility. In order to satisfy local requirements and to offer optimal integration with partner institute resources beyond INSDC, submission and presentation tools are developed and maintained independently at the partner institutes. These tools are available at: http://www.ddbj.nig.ac.jp/, http://www.ebi.ac.uk/ena/ and http://www.ncbi.nlm.nih.gov/ for DDBJ, ENA and NCBI, respectively, and linked from http://www.insdc.org/.
Clear principles on data ownership have been developed by the INSDC databases. Importantly, INSDC databases are data hosts and not owners; while there are certain syntactic and semantic compliance validations for incoming data, data ownership, and hence editorial control of the scientific content, remains with the original data provider. Furthermore, only data owners and their approved delegates are permitted to update their records. Data submitted to one of the partners undergo updates that are mediated by the recipient INSDC institute; i.e. the recipient institute takes permanent responsibility for the interaction between the submitter and the INSDC over any given record or set of records.
Clearly, while such a system brings impartiality, it also leaves scientific quality control at the hands of data providers who, on occasion, are unable to support ongoing updates to their records over extended periods, typically as a result of a change in the focus of the submitter’s laboratory or as a result of employees leaving the domain of research in question. As a primary archive, it is important that INSDC places as few barriers to data providers as possible in order that their data and interpretations are fully disseminated as part of the scientific record. For this reason, INSDC content spans many levels of completeness, thoroughness and, ultimately, reliability as a feed of information into an analysis. Recognizing this issue, it is the policy of the INSDC databases to strive for systems under which quality, completeness and thoroughness can be evaluated and expressed to allow users to make best judgements on confidence in different INSDC records under different analyses.
While it is mainstream dogma in bioinformatics that any paper in which new sequence is described cites INSDC accession numbers that are associated with sequences that have been submitted by the authors of the paper, this ‘mandatory submission’ concept arose not passively, but through the efforts of INSDC member institutions and other proponents of open data sharing. As an example of good practice in public data dissemination, INSDC partners acknowledge the ongoing support of the publishers of major life science journals in this endeavour.
INSDC data are provided openly and free of charge to users. While many records are made publicly available immediately following submission, those kept confidential prior to publication are released publicly as soon as the work is presented in a publication. It is necessary, in order to comply with the consent agreements of human donors who have provided material for sequencing, to require authorization over access to some records; INSDC institutes work under their respective legislative systems with the appropriate ethical bodies and committees to achieve appropriate levels of security.
The INSDC has a long established International Advisory Committee that is charged with providing INSDC with scientific and strategic advice on development and policy issues (see http://www.insdc.org/advisors.html). The senior scientists who make up the committee also play an important role as advocates for the INSDC [(5) and http://www.insdc.org/documents/open_letter.txt].
CONTENT IN 2010
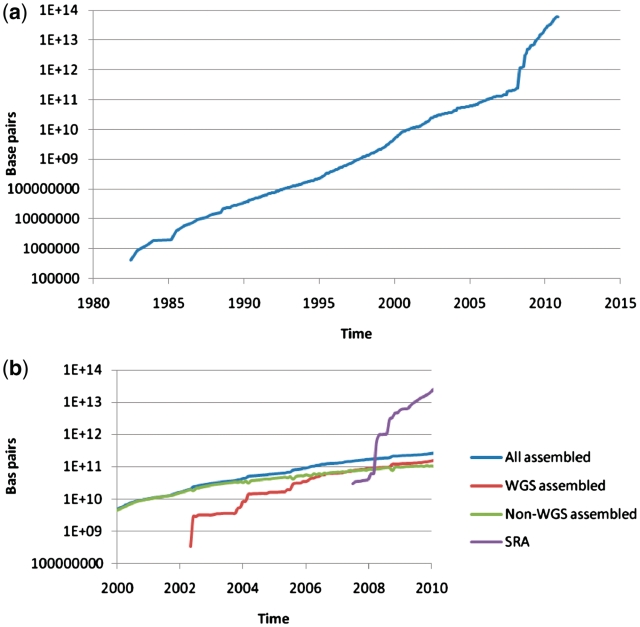
In 2010, INSDC databases have grown overall around 3-fold in terms of the number of bases (Figure 1). Behind this absolute growth are increases in the numbers of assembled sequences of 19% (from 164 to 195-million sequences) and a greater than two-fold increase in the number of next-generation-based experiments in SRA (from around 13 000 to 31 000). While it is not surprising that raw next-generation sequencing data contributes the greatest component of data growth in INSDC databases, a slight but persistent pattern of falling rate of accumulation of assembled sequences is evident. While the causes of this are unclear, amongst the many possible explanations are the trend towards less complete (in traditional terms) genome sequencing, and hence a lower likelihood of a need of data generators to present conventional assembled sequence and functional annotation to the public; indeed it is clear from a breakdown of assembled sequence base contributions (Figure 1b) that assembled sequence records from whole genome shotgun studies now contribute an increasingly significant component of assembled sequences overall. Further explanations, perhaps, include the saturation of sequencing capacity by next-generation sequencing machines whose outputs remain to date, in comparison with those of capillary electrophoresis platforms, less amenable to sequence assembly methods.
Figure 1.
(a) Base pairs in INSDC over time, excluding the Trace Archive (raw data from capillary sequencing platforms). Cumulative data volume in base pairs over time. (b) Base pairs in INSDC over time since 2002, broken down into selected data components. Cumulative data volume in base pairs broken down into assembled sequence (whole genome shotgun methods and others) and raw next-generation-sequence data.
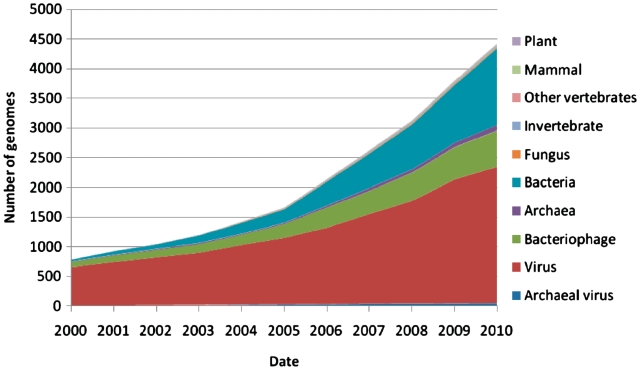
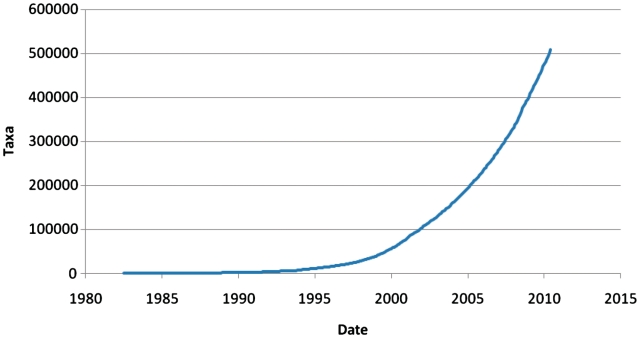
Despite this slowing of growth in assembled sequence submissions to INSDC databases, it is clear that the catalogue of public domain genomes continues to grow rapidly (Figure 2). Furthermore, while the taxonomic diversity of complete genomes has grown with increasing rate over time, it is clear that overall taxonomic coverage, albeit for many taxa with very sparse sequence representation, has also undergone increasing growth over time (Figure 3).
Figure 2.
Growth in complete genomes. The layered chart shows the number of complete genomes available from INSDC databases over time. The end of 2010 time point is conservatively (linearly) extrapolated from October 2010 figures, which are the latest available at the time of submission.
Figure 3.
Taxonomic coverage. Growth in the number of taxa with associated sequence (or with subordinate taxa with associated sequence) over time.
FUTURE CHALLENGES
Throughout the history of nucleic acid sequencing, experimentalists and data resource providers have become accustomed to exponential growth in data (Figure 1), resulting from both increased automation and broadening adoption of molecular methods into the mainstream life sciences. While this traditional exponential growth has created challenges at technical, social and economic levels, the recent technology shift into ‘next-generation’ sequencing platforms has brought a step-change to the nature of growth, which provides perhaps the greatest challenge to date for INSDC partners.
Next-generation technologies bring new technical challenges. Leverage of recent advances in micro-fluidics and imaging technology provide the current next-generation platforms, namely Roche/454, Illumina and Life Technologies/SOLiD, with the capacity to yield greatly increased numbers of parallel reads from clonally amplified single molecules. So-called ‘next next-generation’ platforms promise to bring this level of parallel output to direct single molecule sequencing through the use of advanced imaging and other sensor technologies. While the level of parallel output brings in itself an impressive step-change in data volumes, it is the unprecedented growth in throughput that we have seen to date that is perhaps more challenging. In extreme cases, we have witnessed aggressive technology improvements for a next-generation platform with yield doubling time of as little as 5 months. When contrasted with the historical doubling time for, say, disk density per unit cost, of ∼18 months, it is clear that simply containing next-generation data in affordable storage media, under a sustainable economic model, is a key technical challenge. As stakeholders, the INSDC partners are contributing to and engaging with local and community efforts to develop data reduction, compression and other methodologies to rise to the challenge of aggressive sequencing technology growth. One part of this activity drives algorithmic development and method optimization, while another asks critical questions as to the value of the different data components that we preserve within our archives.
The advent of next-generation sequencing technology brings a further challenge, for which likely solutions will come less from cutting-edge technical developments, but rather from social and organizational innovation. The enormous reduction in cost has driven a broad uptake of sequencing across a huge breadth of applications well beyond traditional genome and transcriptome sequencing for the purposes of functional annotation. Through the availability of new approaches such as next-generation sequence-based epigenomics, resequencing, metatranscriptomics and quantitative expression, sequencing has now become a staple general assay platform for the life scientist. Furthermore, in many cases, it is no longer necessary even for the user to understand fully the intricacies of the sequencing-based components of their work. How, then, can the INSDC continue to serve best such a broadening user base? Developing expertise in all new sequencing related applications within INSDC through training and new recruitment is unlikely to be sustainable. The strategy adopted by INSDC has been the establishment of close collaborations with groups having specialized expertise, such as GEO (6) and ArrayExpress (7) for functional genomics experiments. Increasingly in these arrangements, access to INSDC data is provided through these collaborating groups which are better placed for providing specialized data submission, presentation and integration tools. In a further example, work with the Consortium for the Barcoding of Life and the Genomics Standards Consortium has led to an INSDC-provided keyword applied to those records that reach compliance with such standards as the barcode data standard and the community-developed Minimal Information about a Genome Sequence (MIGS) standard [http://www.barcoding.si.edu/PDF/DWG_data_standards-Final.pdf, (8)]. This model is sustainable not only because it apportions work appropriately (generic bioinformatics and data infrastructure work goes to INSDC and work in specialist biological domains goes only to the appropriate experts), but also because it apportions responsibility for justification to funders and compliance with governance protocols to those most equipped for these tasks—the experts in the domain.
FUNDING
EBI by the European Molecular Biology Laboratory; the European Commission; Wellcome Trust, at DDBJ by the Ministry of Education, Culture, Sports, Science and Technology of Japan; at the NCBI by the Intramural Research Program of the National Institutes of Health; National Library of Medicine. Funding for open access charge: European Molecular Biology Laboratory.
Conflict of interest statement. None declared.
REFERENCES
- 1.Leinonen R, Sugawara H, Shumway M on behalf of the International Nucleotide Sequence Database Collaboration. The Sequence Read Archive. Nucleic Acids Res. 2011;39:D19–D21. doi: 10.1093/nar/gkq1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kaminuma E, Mashima J, Kodama Y, Gojobori T, Ogasawara O, Okubo K, Takagi T, Nakamura Y. DDBJ launches a new archive database with analytical tools for next-generation sequence data. Nucleic Acids Res. 2010;38:D33–D38. doi: 10.1093/nar/gkp847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Leinonen R, Birney E, Bower L, Cerdeno-Tárraga A, Cheng Y, Cleland I, Faruque N, Goodgame N, Gibson R, Jang M, et al. The European Nucleotide Archive. Nucleic Acids Res. 2011;39:D28–D31. doi: 10.1093/nar/gkq967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Brunak S, Danchin A, Hattori M, Nakamura H, Shinozaki K, Matise T, Preuss D. Nucleotide Sequence Database Policies. Science. 2002;298:1333. doi: 10.1126/science.298.5597.1333b. [DOI] [PubMed] [Google Scholar]
- 6.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update–from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37(Suppl. 1):D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Field D, Garrity G, Gray T, Morrison N, Selengut J, Sterk P, Tatusova T, Thomson N, Allen MJ, Angiuoli SV, et al. The minimum information about a genome sequence (MIGS) specification. Nature Biotechnol. 2008;26:541–547. doi: 10.1038/nbt1360. [DOI] [PMC free article] [PubMed] [Google Scholar]