Abstract
ConsensusPathDB is a meta-database that integrates different types of functional interactions from heterogeneous interaction data resources. Physical protein interactions, metabolic and signaling reactions and gene regulatory interactions are integrated in a seamless functional association network that simultaneously describes multiple functional aspects of genes, proteins, complexes, metabolites, etc. With 155 432 human, 194 480 yeast and 13 648 mouse complex functional interactions (originating from 18 databases on human and eight databases on yeast and mouse interactions each), ConsensusPathDB currently constitutes the most comprehensive publicly available interaction repository for these species. The Web interface at http://cpdb.molgen.mpg.de offers different ways of utilizing these integrated interaction data, in particular with tools for visualization, analysis and interpretation of high-throughput expression data in the light of functional interactions and biological pathways.
INTRODUCTION
Knowledge of the functional interactions between physical entities in the cell has high explanatory power regarding biological processes in health and disease (1). Thus, numerous methods for mapping functional association networks such as physical protein interaction networks, metabolic and signaling pathways and gene regulatory networks have been applied in many organisms. The data resulting from such analyses are currently interspersed in hundreds of databases that typically contain only a single aspect of functional interactions of genes, proteins, etc. (2). For example, some databases are specialized on storing protein–protein interaction data, while others focus on the curation of biochemical pathways and still others on gene regulatory interactions. In the cell, however, all different types of functional interactions are operative at the same time: to give an example scenario, genes are regulated to produce proteins that interact physically with other proteins to form complexes that catalyze metabolic reactions. ConsensusPathDB, which we previously reported in (3), assembles a functional association network from multiple heterogeneous public interaction resources by integrating physical entities based on their accession numbers and functional interactions based on their participants. As the combined interaction network in ConsensusPathDB reveals multiple functional aspects of cellular entities at the same time by combining highly complementary data, it is closer to biological reality than the separate source networks. The content of ConsensusPathDB can be exploited in different ways and contexts through its public Web interface at http://cpdb.molgen.mpg.de. It features interaction querying and visualization, network validation and several tools for the interaction- and pathway-level interpretation of user-specified gene or protein expression data.
In this database update report, we highlight the major extensions of ConsensusPathDB regarding database content and functionality of its Web interface.
DATABASE CONTENT: NEW SOURCE DATABASES, NEW INTERACTIONS AND NEW TAXONOMIC SPECIES
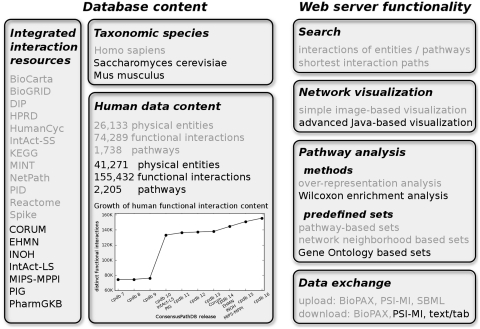
Since the previous database report (3), the human interaction content of ConsensusPathDB has been increased significantly (Figure 1, left panel). Due to the integration of six additional interaction data resources and updates on the previously integrated 12 resources, the human interaction data in ConsensusPathDB have more than doubled from 74 289 to 155 432 unique complex functional interactions. The newly integrated data include complex protein interactions from Corum (4), large-scale protein interaction networks from IntAct (5) (designated IntAct-LS), manually curated protein–protein interactions from MIPS-MPPI (6), protein–protein interactions from the Pathogen Interaction Gateway (PIG) meta-database (7), the Edinburgh Human Metabolic Network reconstruction (EHMN) (8) and biological pathways from INOH (http://www.inoh.org). We have additionally imported 5238 physical interactions between human transcription factors published recently in ref. 9. Furthermore, pathway definitions in the form of lists of genes participating in biological pathways were imported from PharmGKB (10) for use in pathway-based analysis of expression data. With the addition of PIG, 20 098 host–pathogenic protein–protein interactions were introduced into ConsensusPathDB involving proteins from 864 viral and bacterial species. Thus, the integrated ConsensusPathDB network can now additionally serve as explanatory basis in the context of infectious diseases.
Figure 1.
ConsensusPathDB content and Web interface functionality. Content and features that have been described in our previous database report (3) are displayed in gray font, new items in black. The plot in the left panel shows the growth of the human interaction data in ConsensusPathDB since the last database report (ConsensusPathDB release 7, 74 289 interactions) through the releases until present (release 16, 155 432 interactions). Source database additions that contribute interactions to ConsensusPathDB are listed under the corresponding release in the plot.
Table 1 shows the number of human interactions imported from each database, as well as the pairwise overlaps of source databases. To assess these overlaps and to avoid redundant interactions in ConsensusPathDB, physical entities and functional interactions from source databases are mapped to each other. The mapping process is detailed in Supplementary Data.
Table 1.
Pairwise overlaps between human interaction databases in terms of shared functional interactions as of September 2010
| Reactome | Kegg | Humancyc | Pid | Biocarta | Netpath | Inoh | Ehmn | Intact-ss | Intact-ls | Dip | Mint | Hprd | Spike | Biogrid | Pig | Corum | Mips-mppi | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Reactome | 5953 | 286 | 147 | 157 | 85 | 53 | 199 | 327 | 152 | 19 | 69 | 73 | 387 | 125 | 233 | 489 | 108 | 8 |
| Kegg | 286 | 1747 | 242 | 0 | 4 | 0 | 382 | 1135 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| Humancyc | 147 | 242 | 1765 | 1 | 4 | 0 | 122 | 254 | 6 | 2 | 3 | 4 | 21 | 2 | 11 | 33 | 6 | 0 |
| Pid | 157 | 0 | 1 | 5664 | 346 | 257 | 94 | 0 | 129 | 12 | 109 | 106 | 427 | 257 | 351 | 520 | 95 | 15 |
| Biocarta | 85 | 4 | 4 | 346 | 2260 | 129 | 51 | 8 | 62 | 7 | 41 | 39 | 110 | 173 | 114 | 141 | 40 | 7 |
| Netpath | 53 | 0 | 0 | 257 | 129 | 2139 | 63 | 0 | 262 | 13 | 201 | 277 | 1304 | 475 | 694 | 1121 | 33 | 19 |
| Inoh | 199 | 382 | 122 | 94 | 51 | 63 | 2346 | 395 | 30 | 5 | 29 | 28 | 75 | 54 | 71 | 98 | 23 | 1 |
| Ehmn | 327 | 1135 | 254 | 0 | 8 | 0 | 395 | 3873 | 0 | 0 | 0 | 0 | 0 | 4 | 0 | 0 | 0 | 0 |
| Intact-ss | 152 | 0 | 6 | 129 | 62 | 262 | 30 | 0 | 12 352 | 416 | 427 | 3757 | 3743 | 4681 | 2892 | 6303 | 201 | 65 |
| Intact-ls | 19 | 0 | 2 | 12 | 7 | 13 | 5 | 0 | 416 | 12 215 | 36 | 3798 | 5152 | 8592 | 6334 | 10 836 | 16 | 12 |
| Dip | 69 | 0 | 3 | 109 | 41 | 201 | 29 | 0 | 427 | 36 | 1701 | 378 | 842 | 522 | 736 | 1286 | 64 | 29 |
| Mint | 73 | 0 | 4 | 106 | 39 | 277 | 28 | 0 | 3757 | 3798 | 378 | 13 994 | 6766 | 5893 | 4928 | 11 834 | 99 | 50 |
| Hprd | 387 | 0 | 21 | 427 | 110 | 1304 | 75 | 0 | 3743 | 5152 | 842 | 6766 | 404 10 | 112 38 | 158 68 | 318 76 | 468 | 249 |
| Spike | 125 | 0 | 2 | 257 | 173 | 475 | 54 | 4 | 4681 | 8592 | 522 | 5893 | 112 38 | 222 32 | 9975 | 181 00 | 140 | 113 |
| Biogrid | 233 | 0 | 11 | 351 | 114 | 694 | 71 | 0 | 2892 | 6334 | 736 | 4928 | 158 68 | 9975 | 334 35 | 193 15 | 261 | 195 |
| Pig | 489 | 0 | 33 | 520 | 141 | 1121 | 98 | 0 | 6303 | 108 36 | 12 86 | 118 34 | 318 76 | 181 00 | 193 15 | 100 459 | 372 | 302 |
| Corum | 108 | 0 | 6 | 95 | 40 | 33 | 23 | 0 | 201 | 16 | 64 | 99 | 468 | 140 | 261 | 372 | 1667 | 9 |
| Mips-mppi | 8 | 0 | 0 | 15 | 7 | 19 | 1 | 0 | 65 | 12 | 29 | 50 | 249 | 113 | 195 | 302 | 9 | 323 |
The numbers in the main diagonal denote the distinct interactions imported from each database.
Apart from extending the human functional interaction network, we have created ConsensusPathDB instances for two more organisms: Saccharomyces cerevisiae and Mus musculus, integrating eight interaction resources each: Reactome (11), KEGG (12), BioCyc (13), IntAct (5), DIP (14), MINT (15), BioGRID (16) and MIPS (6,17). The mouse instance additionally includes 1145 interactions between mouse transcription factors obtained from ref. 9. As in the case of the human ConsensusPathDB instance, only metabolic reactions have been imported from KEGG in the mouse and yeast database instances. This is due to the fact that signaling reactions are not made available by KEGG in any computer-readable format. However, KEGG’s signaling pathways are stored in ConsensusPathDB in the form of gene lists for use in the context of gene expression analyses described below.
Overall, ConsensusPathDB currently contains 41 271 physical entities, 155 432 functional interactions and 2205 biological pathways in human; 14 532 physical entities, 194 480 functional interactions and 734 biological pathways in yeast; and 21 946 physical entities, 13 648 functional interactions and 1381 biological pathways in mouse. The numbers correspond to the content after integration, i.e. unique item counts (for example, the number of non-unique human interactions before integration is 306 003). Our meta-database is updated every 3 months with the newest releases of its interaction resources.
For the vast majority of functional interactions and physical entities, annotation in the form of literature references and sequence database identifiers, respectively, is imported from the source databases. Literature references are especially useful for protein–protein interactions, as they often serve for interaction confidence estimations. We do not make any judgments on the quality of interactions: all interactions from all source databases are treated equally. For example, physical interactions detected by both large-scale and small-scale experiments are accommodated in ConsensusPathDB without applying any interaction filtering. The ConsensusPathDB users can themselves opt to use filtering based, e.g. on the number of publications, the scale of the interaction detection method or the number of source databases per interaction, since this information is stored and provided in ConsensusPathDB.
For all physical entities, interactions and pathways, the different source databases are recorded and links to the original data are provided where applicable.
NEW WEB SERVER FUNCTIONALITY
Apart from boosting the interaction content in the ConsensusPathDB repository, we have further developed its publicly accessible Web interface (http://cpdb.molgen.mpg.de) to add new functionality (Figure 1, right panel). This includes mainly (i) an advanced, more flexible network visualization framework that features for example overlay of expression values on physical entity nodes and (ii) new facilities in the gene expression data analysis context for the detection of interaction sub-networks and other functional gene groups that have changed activity between phenotypes.
Network visualization
After searching for interactions of particular physical entities, biological pathways or shortest paths of interactions connecting any two physical entities from ConsensusPathDB, the Web interface user has presently two choices for visualizing selected interactions. These choices include the previously described, static-image-based visualization framework and a new, Java-applet-based framework. Both frameworks display interaction networks in the same style, so switching between them involves hardly any user acclimatization. While the latter framework requires a Java Runtime Environment to be installed on the client computer and thus has higher processor and workspace requirements than a simple computer image, it has several advantages, especially when it comes to visualizing larger networks. Network nodes (physical entities/functional interactions) are movable and can be rearranged automatically using different layout methods. Network viewing is further facilitated through the zoom function controlled by the computer mouse wheel. In this Java-based visualization environment, gene/protein expression data can be overlaid on the nodes of a currently viewed network to enable the interaction network-based interpretation of these data (Figure 2).
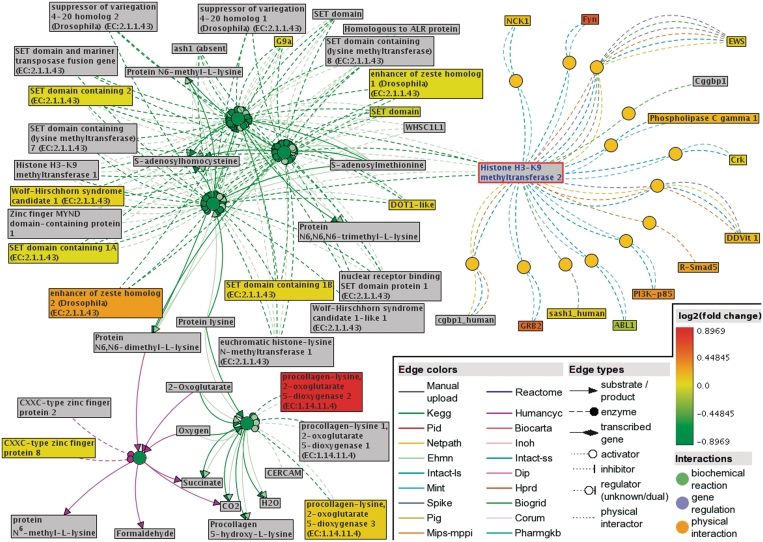
Figure 2.
A functional interaction sub-network visualized by ConsensusPathDB’s new network visualization framework. This sub-network defines the neighborhood-based entity set centered by SUV39H2 (Histone H3-K9 methyltransferase 2, highlighted with red frame in the network) and containing its direct physical interactors, as well as enzymes of neighboring biochemical reactions. The network consists of 13 physical interactions (orange circles) and five biochemical reactions (green circles) from nine different databases (interaction origins are encoded as edge colors). Gene expression data by Yu et al. (20) are overlaid as log2(fold change) values on the physical entity nodes (rectangles). Protein products of measured genes are colored according to the fold expression change (see legend), the rest of the physical entities in the network are gray. Based on the Yu et al. data, the Wilcoxon enrichment analysis tool of ConsensusPathDB suggests that members of this neighborhood-based entity set show jointly significant increase in transcription levels in metastatic prostate cancer compared to primary prostate carcinoma.
Network- and pathway-based analysis of gene expression data
Using the Web interface of ConsensusPathDB, gene expression data can be analyzed with statistical methods on the level of predefined functional gene sets. These gene sets are based on neighborhood in the functional interaction network, cooperation in curated biochemical pathways or, since recently, co-annotation with Gene Ontology (18) categories. One possibility to interpret the gene expression data is through gene set over-representation analysis—a functionality that we have described in our previous database report (3). Here, the user uploads a list of genes that are differentially expressed in a phenotype of interest, typically a disease phenotype, compared to a control phenotype. Based on the hypergeometric test, predefined functional gene sets such as pathways or interaction sub-networks are identified that contain significantly many of the uploaded genes of interest. For example, if differentially expressed genes are over-represented in a network region, this can be an indicator that this region may be dysregulated in the phenotype in question. In addition to over-representation analysis, we have implemented a gene set enrichment analysis method, which we have reported in (19). In this approach, denoted Wilcoxon enrichment analysis, the complete set of measured genes is uploaded with two expression values per gene, rather than just a non-weighted list of genes that pass a significance threshold as in the case of over-representation analysis. The per-gene values typically represent gene expression levels in the two phenotypes being compared. For every predefined gene set, a Wilcoxon signed-rank test is calculated to evaluate the joint expression difference of the whole predefined gene set rather than individual genes. In other words, even if a predefined functional gene set, such as a pathway, contains no genes with significant differential expression, the joint expression of the group of genes may be significantly changed, indicating potential pathway deregulation on a low but nonetheless consistent gene level.
Use case: functional network-based analysis of gene expression data
To demonstrate the utility of the new Wilcoxon enrichment analysis functionality and the overlay of expression values on interaction networks within the new Java network visualization environment, we applied these tools on gene expression measurements from Yu et al. (20) comparing prostate carcinoma against metastatic prostate cancer patients. The Yu et al. data were downloaded from Oncomine 3.0 (21) in February 2009. These data constitute gene expression measurements in 64 prostate carcinoma samples and 25 metastatic prostate cancer samples and are summarized in Oncomine in the form of mean normalized gene expression values for the two sample cohorts, as well as a t-test P-value reflecting the significance of differential gene expression. We additionally filtered the data to exclude ESTs and ambiguously identified genes. For Wilcoxon enrichment analysis in ConsensusPathDB, we uploaded the resulting list of 7807 genes together with their mean expression values for both patient cohorts. We selected interaction neighborhood-based entity sets of radius 1, curated pathways, and Gene Ontology level 3 biological process categories as predefined functional sets for enrichment analysis with default parameter settings. Results, summarized in Supplementary Table S1, clearly correspond to the hallmarks of human cancer (22): changes in the cell cycle, transcription, translation, signaling, angiogenesis and immune response. For example, among the pathways whose activity is significantly changed in metastatic cancer compared to primary carcinoma according to the Wilcoxon enrichment analysis (Supplementary Table S1) are ‘Ribosome’ (KEGG) [see (23)]; ‘Translation’ (Reactome); ‘Mitotic cell cycle’ (Reactome); ‘Interleukin-5 immune pathway’ (NetPath); ‘VEGF, hypoxia and angiogenesis’ (BioCarta); as well as several cancer-related signaling pathways like ‘GPCR signaling’ (Reactome); ‘PDGFR-beta signaling’ (Pathway Interaction Database); ‘ERK signaling’ (Reactome); ‘RAS signaling’ (Reactome); ‘JAK/STAT signaling’ (INOH). Notably, KEGG’s ‘Non-small cell lung cancer pathway’ is also among the significantly enriched pathways. Although no Gene Ontology categories were significant at the 0.05 P-value threshold after correction for multiple testing, among the categories with significant Wilcoxon enrichment P-values (Supplementary Table S1) are ‘Chromosome organization’, ‘Chromatin assembly’, ‘Regulation of organ growth’ and ‘Mitotic cell cycle’.
As for enriched neighborhood-based entity sets (Supplementary Table S1), the most significantly enriched one (Wilcoxon signed-rank test P-value = 8.34e-6) has Histone H3-K9 methyltransferase 2 (gene symbol SUV39H2) as the set center. This neighborhood-based entity set is constructed from physical interactions and biochemical reactions originating from overall nine different source databases. The central gene SUV39H2 plays a crucial role in cell cycle, transcriptional regulation and cell differentiation [Gene Ontology annotation, UniProt (24) keywords] and its mutations have been shown to increase the risk of cancer in human and in mouse models (25,26). It is important to mention that SUV39H2 itself is not contained in the expression data set that we uploaded and used for Wilcoxon enrichment analysis, but many of its network neighbors have expression measurements that show coherent transcriptional dysregulation. Figure 2 shows the functional interaction neighborhood sub-network of SUV39H2, where the Yu et al. data are overlaid on protein nodes as logarithmized gene expression fold change. To conclude, our results support previous findings (27) that utilizing functional interaction data can substantially improve expression data interpretation and can point to disease genes that do not come up on the gene expression analysis level alone. Moreover, our results underline the importance of data integration as the SUV39H2-centered sub-network comprises different types of functional interactions originating from nine interaction databases.
Examples for further significantly enriched neighborhood-based entity sets include the ones centered by ribosomal proteins (e.g., 40S ribosomal protein S4, Y isoform 2: UniProt: RS4Y2_HUMAN), Nucleosome assembly complex protein 1-like 4 (UniProt: NP14L_HUMAN), cell cycle proteins (e.g., MAT1 and MAP kinase p38 delta: UniProt: MK13_HUMAN) and by the transcription factor SP1 that, according to UniProt annotation, may modulate the cellular response to DNA damage.
AVAILABILITY
The Web interface to ConsensusPathDB is freely available to academic users through http://cpdb.molgen.mpg.de.
The protein interaction part of the ConsensusPathDB interaction network can be downloaded in tab-delimited or PSI-MI (28) formats. While the complete database content is not downloadable due to licensing limitations imposed by several source databases, we provide a list of identifiers of matching interactions across source databases upon request. Furthermore, we offer web services for automated pathway analysis of gene expression data. Through the ConsensusPathDB plugin (29) for the Cytoscape network visualization and analysis software (30), experimentalists can automatically mine evidences for sets of newly detected protein interactions from ConsensusPathDB and highlight novel interactions among them. For the interaction data, web services, and Cytoscape plugin, please visit the ‘data access’ section of ConsensusPathDB’s Web site.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Max Planck Society (IMPRS-CBSC); the European Commission under its 7th Framework Programme with the grant APO-SYS (Health-F4-2007-200767); the German Ministry for Research with the grant PREDICT (0315428A); and the Austrian Nationalstiftung and the Austria Wirtschaftsservice GmbH in the framework of the IMGuS research program. Funding for open access charge: European Union’s project APO-SYS (Health-F4-2007-200767).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank the developers of all ConsensusPathDB’s source databases for making their interaction data available.
REFERENCES
- 1.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Bader GD, Cary MP, Sander C. Pathguide: a pathway resource list. Nucleic Acids Res. 2006;34:D504–D506. doi: 10.1093/nar/gkj126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kamburov A, Wierling C, Lehrach H, Herwig R. ConsensusPathDB – a database for integrating human functional interaction networks. Nucleic Acids Res. 2009;37:D623–D628. doi: 10.1093/nar/gkn698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Ruepp A, Waegele B, Lechner M, Brauner B, Dunger-Kaltenbach I, Fobo G, Frishman G, Montrone C, Mewes H. CORUM: the comprehensive resource of mammalian protein complexes – 2009. Nucleic Acids Res. 2010;38:D497–D501. doi: 10.1093/nar/gkp914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Aranda B, Khadake J, Kerssemakers J, Leroy C, Menden M, Michaut M, Montecchi-Palazzi L, Neuhauser SN, Orchard S, Perreau V, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mewes H, Ruepp A, Frishman D, Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, et al. The MIPS mammalian protein–protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 7.Driscoll T, Dyer MD, Murali TM, Sobral BW. PIG – the pathogen interaction gateway. Nucleic Acids Res. 2009;37:D647–D650. doi: 10.1093/nar/gkn799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ma H, Sorokin A, Mazein A, Selkov A, Selkov E, Demin O, Goryanin I. The Edinburgh human metabolic network reconstruction and its functional analysis. Mol. Syst. Biol. 2007;3:135. doi: 10.1038/msb4100177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ravasi T, Bertin N, Carninci P, Daub CO, Forrest ARR, Gough J, Grimmond S, Han J, Hashimoto T, Hide W, et al. An atlas of combinatorial transcriptional regulation in mouse and man. Cell. 2010;140:744–752. doi: 10.1016/j.cell.2010.01.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Thorn CF, Klein TE, Altman RB. Pharmacogenomics and bioinformatics: PharmGKB. Pharmacogenomics. 2010;11:501–505. doi: 10.2217/pgs.10.15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews L, Jassal B, Kanapin A, Lewis S, Mahajan S, May B, Schmidt E, Vastrik I, Wu G, Birney E, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.López-Bigas N, Karp PD, Ouzounis CA, Moore-Kochlacs C, Goldovsky L, Kaipa P, Ahrén D, Tsoka S, Darzentas N, Kunin V. Expansion of the BioCyc collection of pathway/genome databases to 160 genomes. Nucleic Acids Res. 2005;33:6083–6089. doi: 10.1093/nar/gki892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wood V, Dolinski K, Tyers M, Breitkreutz B, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Güldener U, Münsterkötter M, Oesterheld M, Pagel P, Ruepp A, Mewes H, Stümpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makrantonaki E, Adjaye J, Herwig R, Brink TC, Groth D, Hultschig C, Lehrach H, Zouboulis CC. Age-specific hormonal decline is accompanied by transcriptional changes in human sebocytes in vitro. Aging Cell. 2006;5:331–344. doi: 10.1111/j.1474-9726.2006.00223.x. [DOI] [PubMed] [Google Scholar]
- 20.Yu YP, Landsittel D, Jing L, Nelson J, Ren B, Liu L, McDonald C, Thomas R, Dhir R, Finkelstein S, et al. Gene expression alterations in prostate cancer predicting tumor aggression and preceding development of malignancy. J. Clin. Oncol. 2004;22:2790–2799. doi: 10.1200/JCO.2004.05.158. [DOI] [PubMed] [Google Scholar]
- 21.Rhodes DR, Kalyana-Sundaram S, Mahavisno V, Varambally R, Yu J, Briggs BB, Barrette TR, Anstet MJ, Kincead-Beal C, Kulkarni P, et al. Oncomine 3.0: genes, pathways, and networks in a collection of 18,000 cancer gene expression profiles. Neoplasia. 2007;9:166–180. doi: 10.1593/neo.07112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hanahan D, Weinberg RA. The hallmarks of cancer. Cell. 2000;100:57–70. doi: 10.1016/s0092-8674(00)81683-9. [DOI] [PubMed] [Google Scholar]
- 23.Vaarala MH, Porvari KS, Kyllönen AP, Mustonen MV, Lukkarinen O, Vihko PT. Several genes encoding ribosomal proteins are over-expressed in prostate-cancer cell lines: confirmation of L7a and L37 over-expression in prostate-cancer tissue samples. Int. J. Cancer. 1998;78:27–32. doi: 10.1002/(sici)1097-0215(19980925)78:1<27::aid-ijc6>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 24.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Yoon KA, Hwangbo B, Kim IJ, Park S, Kim HS, Kee HJ, Lee JE, Jang YK, Park JG, Lee JS. Novel polymorphisms in the SUV39H2 histone methyltransferase and the risk of lung cancer. Carcinogenesis. 2006;27:2217–2222. doi: 10.1093/carcin/bgl084. [DOI] [PubMed] [Google Scholar]
- 26.Peters AH, O'Carroll D, Scherthan H, Mechtler K, Sauer S, Schöfer C, Weipoltshammer K, Pagani M, Lachner M, Kohlmaier A, et al. Loss of the Suv39h histone methyltransferases impairs mammalian heterochromatin and genome stability. Cell. 2001;107:323–337. doi: 10.1016/s0092-8674(01)00542-6. [DOI] [PubMed] [Google Scholar]
- 27.Chuang HY, Lee E, Liu YT, Lee D, Ideker T. Network-based classification of breast cancer metastasis. Mol. Syst. Biol. 2007;3:140. doi: 10.1038/msb4100180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sherman D, Tyers M, Salama JJ, Moore S, Ceol A, Chatr-Aryamontri A, Oesterheld M, Stümpflen V, Salwinski L, Nerothin J, et al. Broadening the horizon – level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pentchev K, Ono K, Herwig R, Ideker T, Kamburov A. Evidence mining and novelty assessment of protein–protein interactions with the ConsensusPathDB plugin for Cytoscape. Bioinformatics. 2010;26:2796–2797. doi: 10.1093/bioinformatics/btq522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]