Abstract
The Munich Information Center for Protein Sequences (MIPS at the Helmholtz Center for Environmental Health, Neuherberg, Germany) has many years of experience in providing annotated collections of biological data. Selected data sets of high relevance, such as model genomes, are subjected to careful manual curation, while the bulk of high-throughput data is annotated by automatic means. High-quality reference resources developed in the past and still actively maintained include Saccharomyces cerevisiae, Neurospora crassa and Arabidopsis thaliana genome databases as well as several protein interaction data sets (MPACT, MPPI and CORUM). More recent projects are PhenomiR, the database on microRNA-related phenotypes, and MIPS PlantsDB for integrative and comparative plant genome research. The interlinked resources SIMAP and PEDANT provide homology relationships as well as up-to-date and consistent annotation for 38 000 000 protein sequences. PPLIPS and CCancer are versatile tools for proteomics and functional genomics interfacing to a database of compilations from gene lists extracted from literature. A novel literature-mining tool, EXCERBT, gives access to structured information on classified relations between genes, proteins, phenotypes and diseases extracted from Medline abstracts by semantic analysis. All databases described here, as well as the detailed descriptions of our projects can be accessed through the MIPS WWW server (http://mips.helmholtz-muenchen.de).
INTRODUCTION
The MIPS group has provided biomolecular databases and related resources for more than 20 years (Table 1). To cope with the quest for accuracy, completeness and timeliness, manually curated databases face the problem of ever-growing amounts of data and related information. The dilemma between the effort to maintain databases curated by experts and the available resources is well known but unfortunately persistent. Manual curation has therefore to concentrate on rather specialized subjects. As an alternative, the application of computational tools and the generation of databases of secondary information can provide most of information rather efficiently. While a large number of databases are available, it is difficult to combine and integrate information although the implementation of web services has improved the situation. Therefore, we concentrate along three lines of services: (i) the development and maintenance of primary manually curated databases for a number of specialized areas of interest that are widely used as gold standards, integrating factual information and the biological knowledge as extracted from the literature; (ii) large-scale and comprehensive databases of secondary data such as (1) SIMAP, the exhaustive database of protein similarities, currently containing 38 million non-redundant protein sequences and (2) PEDANT, the comprehensive database of annotated genomes, now containing 3800 genomes; and (iii) EXCERBT, a query system for the retrieval of biological knowledge based on semantic web technology.
Table 1.
URL addresses for MIPS database resources and associated user interfaces
RECENT DEVELOPMENTS
PhenomiR: structured information on microRNA–phenotype relations
Small endogenous non-coding RNA species known as microRNAs (miRNAs) are essential for a wide variety of cellular processes in higher eukaryotes such as cell and organ development. They are directly or indirectly related to a number of diseases, including cancer. In order to collect the ever-growing body of data in a structured and uniform format, we created the PhenomiR database (1).
All information in PhenomiR is extracted from published studies relating miRNAs and diseases or processes, and has been annotated manually to achieve a high quality of the resource. PhenomiR provides a comprehensive annotation of published experimental data and their origin including the mode of miRNA expression (up- or downregulation), the miRNA detection method, cohort information of patient studies and the study design. Assignment of the origin of the samples (patients or type of cell culture) allows analysing whether inhomogeneous results might stem from the differences of the physiology within the sample set. Quantitative levels of miRNA expression are also provided in a readily accessible way by PhenomiR if the data were published by the authors. This information for example allows discrimination between marginally and significantly deregulated miRNAs. The second release of PhenomiR (as of September 2010) contains data from 362 articles that describe 628 experiments. This data set includes 12 189 data points, each representing one deregulated miRNA in an experiment. A survey about the PhenomiR data set reveals that cancers are by far the most thoroughly investigated diseases (76.6%) followed by neurological (6.3%) and cardiovascular (4.7%) disorders.
MiRNA-mediated gene silencing was shown to be involved in a number of cellular processes such as cell growth, cholesterol homeostasis and response to hypoxia. To our knowledge, PhenomiR is the only database that collects altered miRNA expression not only in diseases but also more generally in biological processes. The availability of both kinds of data allows comparing miRNA expression of disease and cellular process in related cell types. Granulocyte development (16 miRNAs) and multiple myeloma (69 miRNAs), for example, share 14 common deregulated miRNAs. All of these miRNAs are upregulated. As both processes are based on cell growth, a similar behaviour in regulatory processes can be expected. The fraction of miRNAs that is specific for multiple myeloma contains a majority (31 of 55) of downregulated miRNAs (Supplementary Figure S1).
MIPSPlantsDB: plant database resource for integrative and comparative plant genome research
Massive generation of plant genomics data asks for in-depth analysis and enables powerful comparative analysis. PlantsDB aims to provide data and information resources for individual plant species building a platform for integrative and comparative plant genome research. PlantsDB is constituted from genome databases for Arabidopsis, Medicago, lotus, rice, maize and tomato, barley, Brachypodium and Sorghum. Complementary data resources for repetitive elements and extensive cross-species comparison are also implemented.
The ongoing projects include both model genomes (Arabidopsis thaliana, Arabidopsis lyrata, Brachypodium distachyon and Medicago truncatula) as well as important crop genomes such as barley, Sorghum, maize or tomato. Besides the need for comprehensive, structured genome and knowledge information, genome for the individual species, detailed and comprehensive comparative analysis asks for the availability of a range of plant genomes that represent a wide spectrum and evolutionary range. In addition, these information resources will help to elucidate genomic elements that have not been discovered so far or have been difficult to detect. Consistent, detailed data and structured information resources are a prerequisite for detailed and in-depth cross-species comparisons and comparative phylogenetic analysis. PlantsDB provides a highly flexible modular database infrastructure for a wide range of plant genomic data. The respective species databases are updated and new data are continuously integrated either through adjustment against external resources or via the MIPS’ participation in a range of plant genome sequencing projects.
While individual organism-related databases provide an important pillar of PlantsDB, the focus of PlantsDB is extending beyond individual genomes. PlantsDB aims to make available resources that are species spanning and address and support specific questions in comparative and integrative plant genomics. Topics and resources circumvent integrated resources for the detection and analysis of repeat elements, comparative views and navigation systems. The PlantsDB resources are complemented by BioMOBY based web-service opportunities that support seamless navigation and combination of services provided by PlantsDB and partner databases worldwide.
Similarity Matrix of Proteins: exhaustive protein similarity matrix
The Similarity Matrix of Proteins (SIMAP) is an exhaustive, up-to-date database of pre-calculated similarities and features of protein sequences (2). In order to cover the publicly accessible sequence space comprehensively, SIMAP includes the sequence data from all major protein sequence databases as well as from important repositories of environmental sequences. Until 2010, the size of SIMAP has continuously increased to a recent number of 38 million non-redundant protein sequences collected from more than 80 million database entries. The resulting similarity network consists of more than 400 billion edges, connecting the proteins at the most sensitive level that can be achieved by pair-wise local protein sequence alignments. These unique data can be accessed via a user-friendly web portal, providing customizable search tools and integrating information from sequence similarity, protein domains and sequence clusters. Alternatively, SIMAP data can be freely retrieved through Web-Services by applications and resources. SIMAP thus not only speeds up traditional sequence and genome analyses based on sequence similarities, but also facilitates network-based approaches exploring the protein sequence space.
PEDANT genome database
The PEDANT genome database is a comprehensive database of automatically annotated genomes (3). The current version of PEDANT contains 3802 publicly available genomes (89 archaea, 1076 bacteria, 132 eukaryotes and 2505 viruses). It provides full coverage of the NCBI’s RefSeq database (4). New RefSeq genomes are imported monthly; already imported genomes get updated regularly. Changes of genetic elements are tracked. Obsolete elements are marked as outdated but are not deleted in order to keep inbound Web links functional.
The underlying database system has been adapted for improved scalability and now uses optimized data types in combination with index and data compression. This enables cross-genome queries and increases retrieval performance.
A new pathway coverage method has been added which allows determining metabolic pathways present in a given genome based on computationally derived EC numbers. Integration with other MIPS databases and external resources has been improved. PEDANT proteins are now directly linked to SIMAP and to GBrowse databases hosted at MIPS (i.e. Ustilago maydis).
Metabolomics@MIPS
The enormous amount of data produced by modern kit-based high-throughput metabolomics experiments poses new challenges regarding their biological interpretation in the context of various sample phenotypes. We currently provide three web-based data-analysis tools for metabolomics. metaP-server for high-throughput targeted metabolomics, MassTRIX, (5) for deep-drilling non-targeted metabolomics and TICL (6) for network-based interpretation of a compound list. All servers provide results that link-out to dedicated metabolomics databases such as KEGG (7), HMDB (8), LIPID MAPS (9) and a hand-curated in-house database with a specific focus on kit-based lipidomics read-outs.
EXCERBT: a database of biological object relations built from biomedical texts
The current databases represent factual information, either as primary data collections or derived information. Information derived from factual observations is to the largest extent buried in the scientific literature. As it is more and more impossible to follow all publications in a certain research area, the automatic analysis of free text and its transformation into structured databases become a necessity. Specialized text mining (TM) systems such as GIN (10) or RLIMS-P (11) have been published. Few systems have a broader scope integrating text resources, relation and entity types.
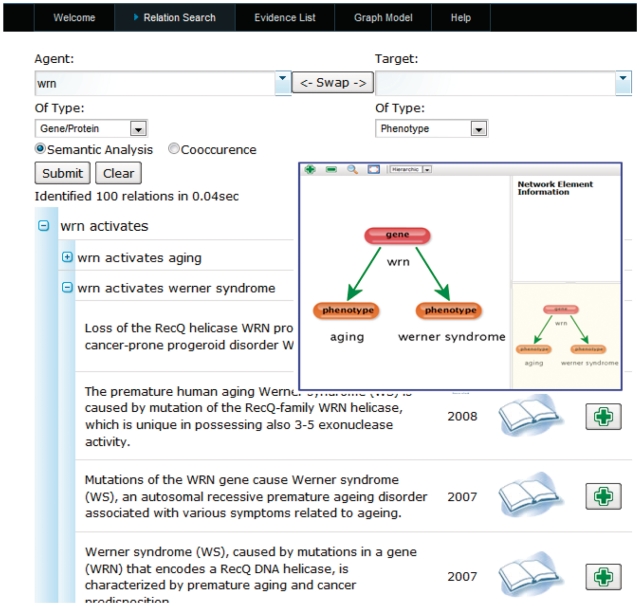
EXCERBT (‘extraction of classified entities and relations from biomedical texts’) is a database of extracted semantic relations from MEDLINE abstracts for the retrieval of a multitude of relation types such as ‘activation’, ‘inhibition’ or ‘phosphorylation’ and a broad range of biomedical entity types. The subject-centric results are presented usually within a few seconds, although the system covers MEDLINE and several smaller sources of literature. Due to the multitude of integrated relation and entity types and the possibility to submit the queries in passive as well as active voice, EXCERBT is a versatile resource to explore the giant combinatorial space between semantically associated biological entities. It contains ∼4 billion relations; EXCERBT is suited to build large-, medium- and small-scale qualitative networks suitable for the systematic exploration of topic-related information (Figure 1).
Figure 1.
EXCERBT interface: (i) “which phenotypes are caused by the wrn gene?”; (ii) list of gene–phenotype pairs and detailed literature evidence; and (iii) graphical representation of selected relations.
PLIPS and CCANCER: a database of gene/protein lists reported in experimental studies in various functional contexts
‘Omics’ technologies provide a spectrum of methods applied in many fields of cellular and molecular biology such as the identification of diagnostic biomarkers, monitoring the effects of drug treatments or profiling transcripts related to onset and progress of diseases. Although very different with respect to the biological system tested, the primary results of the majority of ‘omics’ studies are gene/protein list. Several thousands of independent experimental studies have reported such lists. Although being publicly available, this valuable information was dissolved in hundreds of papers and was not accessible for automatic analysis. To render this information available we collected this type of information by searching through full text papers. We automatically selected tables, which report a list of gene or protein identifiers.
PLIPS (12) is a database of protein lists reported previously by proteomics experimental studies. At the moment PLIPS covers in about 1500 protein lists reported previously by ∼1200 proteomics publications. CCancer (13) is a database of gene lists, which were reported mostly by experimental studies in various biological and clinical settings. At the moment, the database covers 3500 gene lists extracted from 2800 papers published in about 100 peer-reviewed journals. Both databases provide user-friendly computational services. Users can query his/her list of gene/protein identifiers to find a catalogue of previously published studies that report a table of genes/proteins that significantly intersect with a query list.
To understand the functional context of a experimentally identified gene/protein list, MIPS provides a comprehensive collection of online tools for functional profiling. The underlying database covers nearly all available information regarding gene function [Gene Ontology (9;14)], protein interactions [IntAct (15)] and pathway relationships [Reactome (16), KEGG (7)]. The tools implemented robust statistical frameworks and provide computational interfaces for most available public databases. Regarding statistical methodology employed, available web tools can be divided into two categories. The first group [ProfCom (17), PLIPS (12), CCancer (13), GeneSet2MiRNA (18)] employs a modified enrichment analyses schema (19). The second group [KEGG spider (20), R spider (21), PPI spider (22)] implements a novel statistical methodology for the network-based interpretation of a gene list. Finally, GeneSet2MiRNA provides statistical information whether or not a query gene list has a signature of miRNA regulatory activity.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Federal Ministry of Education, Science, Research and Technology (BMBF: GABI Barlex: 0314000C; SysMBo: 0315494A); the European Commission (Framework 6 & 7; Grain Legumes Integrative Project and the Triticeae Genome Project); and the Helmholtz Alliance Systems Biology (CoReNe). Funding for open access charge: Helmholtz Center for Health and Environment.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ruepp A, Kowarsch A, Schmidl D, Buggenthin F, Brauner B, Dunger I, Fobo G, Frishman G, Montrone C, Theis FJ. PhenomiR: a knowledgebase for microRNA expression in diseases and biological processes. Genome Biol. 2010;11:R6. doi: 10.1186/gb-2010-11-1-r6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Rattei T, Tischler P, Arnold R, Hamberger F, Krebs J, Krumsiek J, Wachinger B, Stümpflen V, Mewes W. SIMAP–structuring the network of protein similarities. Nucleic Acids Res. 2008;36:D289–D292. doi: 10.1093/nar/gkm963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Walter MC, Rattei T, Arnold R, Güldener U, Münsterkötter M, Nenova K, Kastenmüller G, Tischler P, Wölling A, Volz A, et al. PEDANT covers all complete RefSeq genomes. Nucleic Acids Res. 2009;37:D408–D411. doi: 10.1093/nar/gkn749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Pruitt KD, Tatusova T, Klimke W, Maglott DR. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Suhre K, Schmitt-Kopplin P. MassTRIX: mass translator into pathways. Nucleic Acids Res. 2008;36:W481–W484. doi: 10.1093/nar/gkn194. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Antonov AV, Dietmann S, Wong P, Mewes HW. TICL–a web tool for network-based interpretation of compound lists inferred by high-throughput metabolomics. FEBS J. 2009;276:2084–2094. doi: 10.1111/j.1742-4658.2009.06943.x. [DOI] [PubMed] [Google Scholar]
- 7.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wishart DS, Knox C, Guo AC, Eisner R, Young N, Gautam B, Hau DD, Psychogios N, Dong E, Bouatra S, et al. HMDB: a knowledgebase for the human metabolome. Nucleic Acids Res. 2009;37:D603–D610. doi: 10.1093/nar/gkn810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fahy E, Sud M, Cotter D, Subramaniam S. LIPID MAPS online tools for lipid research. Nucleic Acids Res. 2007;35:W606–W612. doi: 10.1093/nar/gkm324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ozgur A, Vu T, Erkan G, Radev DR. Identifying gene–disease associations using centrality on a literature mined gene-interaction network. Bioinformatics. 2008;24:i277–i285. doi: 10.1093/bioinformatics/btn182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yuan X, Hu ZZ, Wu HT, Torii M, Narayanaswamy M, Ravikumar KE, Vijay-Shanker K, Wu CH. An online literature mining tool for protein phosphorylation. Bioinformatics. 2006;22:1668–1669. doi: 10.1093/bioinformatics/btl159. [DOI] [PubMed] [Google Scholar]
- 12.Antonov AV, Dietmann S, Wong P, Igor R, Mewes HW. PLIPS, an automatically collected database of protein lists reported by proteomics studies. J. Proteome. Res. 2009;8:1193–1197. doi: 10.1021/pr800804d. [DOI] [PubMed] [Google Scholar]
- 13.Dietmann S, Lee W, Wong P, Rodchenkov I, Antonov AV. CCancer: a bird's eye view on gene lists reported in cancer-related studies. Nucleic Acids Res. 2010;38(Suppl.):W118–W123. doi: 10.1093/nar/gkq515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Aranda B, Achuthan P, am-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de BB, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Antonov AV, Schmidt T, Wang Y, Mewes HW. ProfCom: a web tool for profiling the complex functionality of gene groups identified from high-throughput data. Nucleic Acids Res. 2008;36:W347–W351. doi: 10.1093/nar/gkn239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Antonov AV, Dietmann S, Wong P, Lutter D, Mewes HW. GeneSet2miRNA: finding the signature of cooperative miRNA activities in the gene lists. Nucleic Acids Res. 2009;37:W323–W328. doi: 10.1093/nar/gkp313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Antonov AV, Mewes HW. Complex functionality of gene groups identified from high-throughput data. J. Mol. Biol. 2006;363:289–296. doi: 10.1016/j.jmb.2006.07.062. [DOI] [PubMed] [Google Scholar]
- 20.Antonov AV, Dietmann S, Mewes HW. KEGG spider: interpretation of genomics data in the context of the global gene metabolic network. Genome Biol. 2008;9:R179. doi: 10.1186/gb-2008-9-12-r179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Antonov AV, Schmidt EE, Dietmann S, Krestyaninova M, Hermjakob H. R spider: a network-based analysis of gene lists by combining signaling and metabolic pathways from Reactome and KEGG databases. Nucleic Acids Res. 2010;38(Suppl.):W78–W83. doi: 10.1093/nar/gkq482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Antonov AV, Dietmann S, Rodchenkov I, Mewes HW. PPI spider: a tool for the interpretation of proteomics data in the context of protein–protein interaction networks. Proteomics. 2009;9:2740–2749. doi: 10.1002/pmic.200800612. [DOI] [PubMed] [Google Scholar]