Abstract
The information of protein targets and small molecule has been highly valued by biomedical and pharmaceutical research. Several protein target databases are available online for FDA-approved drugs as well as the promising precursors that have largely facilitated the mechanistic study and subsequent research for drug discovery. However, those related resources regarding to herbal active ingredients, although being unusually valued as a precious resource for new drug development, is rarely found. In this article, a comprehensive and fully curated database for Herb Ingredients’ Targets (HIT, http://lifecenter.sgst.cn/hit/) has been constructed to complement above resources. Those herbal ingredients with protein target information were carefully curated. The molecular target information involves those proteins being directly/indirectly activated/inhibited, protein binders and enzymes whose substrates or products are those compounds. Those up/down regulated genes are also included under the treatment of individual ingredients. In addition, the experimental condition, observed bioactivity and various references are provided as well for user's reference. Derived from more than 3250 literatures, it currently contains 5208 entries about 1301 known protein targets (221 of them are described as direct targets) affected by 586 herbal compounds from more than 1300 reputable Chinese herbs, overlapping with 280 therapeutic targets from Therapeutic Targets Database (TTD), and 445 protein targets from DrugBank corresponding to 1488 drug agents. The database can be queried via keyword search or similarity search. Crosslinks have been made to TTD, DrugBank, KEGG, PDB, Uniprot, Pfam, NCBI, TCM-ID and other databases.
INTRODUCTION
Interaction between small molecule and protein plays a critical role in modulating the intrinsic biological processes. One particular application is the discovery of druggable molecules based on the interaction with the target proteins. Target proteins are often those important ones in the development of specific diseases within the organism. Perturbing their functions by druggable molecules will help to cure the disease or relieve the symptoms. Therefore, the information related to protein targets and small molecule has always been highly valued by biomedical and pharmaceutical sciences. During the last decade, several drug–target interaction databases have been made available online which have largely facilitated the mechanistic study and subsequent research of drug discovery. For instance, Therapeutic Targets Database (TTD) (1) is the first therapeutic target database which sorted known and explored therapeutic proteins and nucleic acid targets and related information for corresponding drugs directed at each of these targets. While another important resource is DrugBank (2) which is a unique database that links detailed drug data to comprehensive drug target information. Such information has lead to integration of further resources and computational methods, such as PDTD (3), TarFisDock (4), STITCH (5) and others (6–9) which have served as valuable platforms for target identification, validation and drug actions.
Herbal ingredients have long been viewed as precious sources by bio-pharmaceutical sciences because of not only the broad chemical structural diversity, but also the wide range of pharmacological activities and comparatively low side effect. It is estimated that approximately one-third (10) of the top-selling drugs in the world are derived from medicinal herbs. A well-known example is the artemisinin from Artemisia annua to treat malaria. In contrast to the well sorted compound–target information for western drugs, similar information for herbal ingredients is rarely found, perhaps partially because of the complicated nature of herbal medicine. To the author's knowledge, only one database (11) mentioned 78 protein targets for 2597 natural compounds, which obviously needs further updating. On the other hand, millions and millions were input to investigate what the potential targets are for promising herbal ingredients with particular pharmaceutical effects, or whether a synthesized compound has similar target profile with any active compounds from herbal plants. As the pharmacological activity could be inferred from related herbs, linking the herbal ingredients to their protein targets may help to bridge information between the natural products and western drugs via protein targets.
Therefore, we here introduced a fully curated database for Herb Ingredients’ Targets (HIT), which is focused on available linking from the single herbal ingredient to its affecting protein targets derived from experimental results. Text mining technologies was firstly applied to PubMed abstracts in order to collect related literatures. Then curation was carefully done to retrieve desired information such as protein target name, action mode, experimental condition and other useful details. As the target information about directly physical interaction for single herbal ingredients is still limited to provide clues to the potential mechanism, indirect targets are collected together as a valuable complement.
THE DATABASE
HIT is currently hosted at http://lifecenter.sgst.cn/hit/. It contains three data fields (Table 1), namely compound information, herb information and protein targets information. The compound information was generated from Chemical Abstracts Service, Pubchem and ‘Dictionary of Natural Products’ (12). TCM-ID (13), a well established TCM integrated resource and the book ‘Traditional Chinese Medicines: Molecular Structures, Natural Sources & Applications’ (14) were used to derive herb information. Considering that more rigorous methods were applied in recent years to detect target–compound interaction, protein targets are curated from Pubmed abstracts published within the last 10 years (2000–2010). The biological annotation for a direct protein target covers detailed action modes of the herbal ingredient, such as activator, inhibitor, binder, agonist, antagonist, substrate or product, and simple target. Kinetic data such as IC50 and Kd/Ki was collected as well if possible. Besides that, the biological effect on indirect targets is indicated as ‘increase/decrease the level of expression/activity’ after being treated with a single herbal compound. The related pathway about the target proteins can be retrieved by following the links to KEGG (15). In addition, the links to TTD and DrugBank could bridge western drugs and herbal molecules at the level of protein targets.
Table 1.
Data fields covered in the entry of HIT
| Compound information | Herb information | Target information |
|---|---|---|
| Generic name | Latin name | Target type (direct/indirect) |
| Structure | Chinese pin yin | Target name |
| IUPAC name | Chinese character | Biological effect |
| Alias | Herb function | IC50 |
| Chemical formula | Kd/Ki | |
| CAS register number | Experimental environment | |
| PubChem CID links | Key description | |
| Code of compound class | Molecular function | |
| Hazard and toxicity | Crosslinks | |
| REICS accession number | Literature support | |
| Other comments |
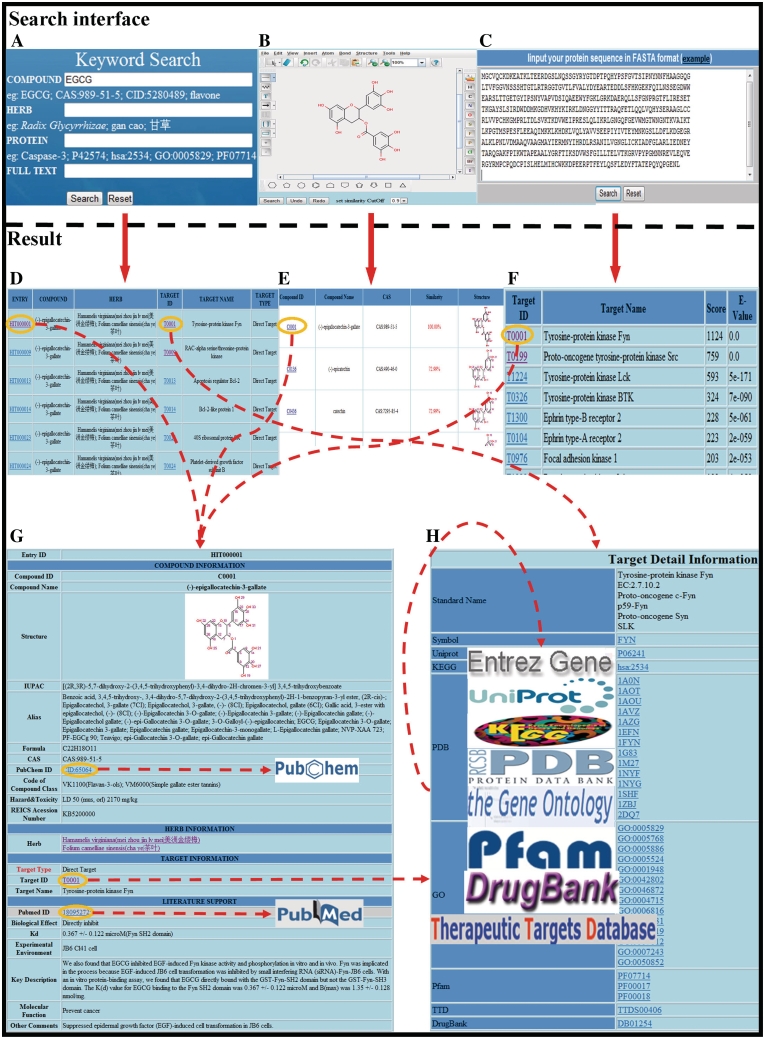
The search interface and results pages are illustrated in Figure 1. HIT can be queried via keyword search or similarity search.
Keyword search can be made via herbal compound information [different names, CAS number, CID number, chemical formula, code of compound class, RTECS Accession Number (http://www.cdc.gov/niosh/rtecs/)], herb information (Latin name, Chinese name as Chinese pinyin or character) or protein target information (various protein/gene name or id). Full text search by keyword is provided as well.
Similarity search is also available via compound structure or protein sequence. The compound structure can be uploaded as a MOL/SDF file or manually drawing with the build-in software MarvinSketch (http://www.chemaxon.com/marvin/). Target similarity search was enabled by Blast program via protein sequence.
Figure 1.
Primary pages in HIT. (A) Screenshot of interface for keyword search. HIT offers three optional search, namely by compound, by herb or by protein target. Text search is also provided by keyword in the whole database. (B) Interface of compound similarity search as an example. (C) Interface of target similarity search via protein sequence. (D) Result page of ‘Keyword Search’ with ‘Compound: EGCG’. (E) Result page of ‘Compound Similarity Search’ with the structure of the compound: EGCG. (F) Result page of ‘Target Similarity Search’ with the sequence of the protein: Fyn kinase (P06241). (G) The further linkage page of the first entry ‘HIT000001’ in D. Variety of chemical information, herbal information and brief protein description is available in this page with crosslinks to NCBI PubChem and PubMed. (H) Screenshot showing the detailed information of protein target: ‘Fyn Kinase’.
METHODS
Herb ingredient names
Herb ingredient names are derived from a well established TCM knowledge database TCM-ID which covers 1102 reputable herbs and 9862 herb ingredients. These compound names were used to screen PubMed abstracts and only those abstracts containing the compound names were recorded.
Keywords library
Establishing a key word library is critically important to retrieve the related literatures. We randomly choose individual compound and checked the full-text review papers to establish this library. Fifty nine keywords are listed in Table 2, which are frequently used to describe the interaction between compound and proteins. The keywords are divided into two types. One is the nouns describing the interaction (Type A), while the other (Type B) is the phrases describing the specific effect such as inhibit the activity of some proteins.
Table 2.
Keyword library to describe the interaction between herbal ingredients and proteins
| Interaction |
Effect | |||
|---|---|---|---|---|
| Positive | Negative | General | ||
| Type A | Agonist; activator | Antagonist; inhibitor | Bind; target; bound | |
| Type B | Activate; Augment; Ameliorate; Derepress; Elevate; Enhance; Hasten; Increase; Induce; Incitate; Initiate Potentiate; Promote; Raise; Stimulate; Up-regulate | Abrogate; Abolish; Against; Attenuate; Antagonize; Block; Blunt; Down regulate; Decrease; Degrade; Diminish; Impair; Inhibit; Reduce; Repress; Suppress | Affect; Interact; Disturb; Regulate; Impact; Influence; Interfere; Modify; Modulate | Activity; Activation; Expression; Level; Pathway; Cleavage; Methylation; Phosphorylation; Severance; Glycosylation; Acetylation |
Text mining and curation
For the above recorded abstracts, text mining was rescanned on them according to below rules:
Rule 1: ‘Compound name’ AND ‘any word in type A’
For instance, the sentence ‘(-)-Epigallocatechin-3-gallate is a novel Hsp90 inhibitor’ matches the rule well.
Rule 2: ‘Compound name’ AND ‘any word in type B interaction’ AND ‘any word in type B effect’
For example, the sentence ‘procyanidin B2 directly inhibited membrane type-1 (MT1)-MMP activity’ is a perfect match.
Manual check was done to all the abstracts being text mined to retrieve useful information into HIT.
Compound similarity search
The calculation of the similarity between two compounds are based on structural fingerprints that generated by Chemistry Development ToolKit (http://almost.cubic.uni-koeln.de/cdk/), using Tanimoto coefficient (16). Given a compound A and a database compound B, the Tanimoto coefficient for binary vectors is defined as
where, a and b are the number of bits set on (‘1’ bits) in molecular fingerprints A and B, respectively and c the number of bits shared by A and B.
This function only accounts for the sum of ‘1’ bits. That is, bits that are set off are not taken into account in similarity calculations. Tanimoto coefficient is typically above 0.8 for similar compounds (17,18).
DISCUSSION AND FUTURE DEVELOPMENT
In summary, HIT is intended to be a primary resource as a complement to other drug–target databases by providing integrative information between medicinal herbs, herb active compounds and the protein target under different experimental conditions. As one important source for drug discovery, some of the herbal ingredients are under intensive pharmacological research, while plenty of them are still to be discovered during which the molecular mechanism is a big challenge. The application of HIT may represent a valuable support to facilitate the mechanistic study of herbal medicine, to discover new druggable molecules, as well as to identify potential therapeutic targets.
However, the action mechanism of herbal medicine is typically featured as ‘multiple ingredients and multiple targets’ which may differ from western drugs to a large extent. The actual biological effects would be much more complicated under different situations when different compounds are grouped together into one herb. It should be aware of that, the biological function a compound A does not always imply the same function for a herb X which contains A because herb X often contains many other compounds. The global and collective effects of many compounds may be different from each single compound. Thus, it is advised that multiple factor analysis and statistical methods should be applied coupled with corresponding experimental and clinical results when efforts are made to drug discovery.
HIT is planned for further enlargement. We will continue collecting target information for more herbal active compounds. Disease condition and batch query function will be considered as well. In addition, HIT is free for academic use. The data can be downloaded upon individual request.
FUNDING
Ministry of Science and Technology, China (2008BAI64B02, 2009ZX10004-601 and 2010CB833601, partial); National Natural Science Foundation of China (30900832 and 30976611); Ministry of Education (NCET-08-0399); Shanghai Municipal Education Commission (08ZZ18); ‘Shu Guang’ project supported by Shanghai Municipal Education Commission and Shanghai Education Development Foundation (07SG22); Shanghai Baiyulan Funding (2010B127).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENT
We thank Dr Guoqing Zhang for helping us to set up the web server.
REFERENCES
- 1.Zhu F, Han BC, Kumar P, Liu XH, Ma XH, Wei XN, Huang L, Guo YF, Han LY, Zheng CJ, et al. Update of TTD: Therapeutic Target Database. Nucleic Acids Res. 2010;38:D787–D791. doi: 10.1093/nar/gkp1014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Gao Z, Li H, Zhang H, Liu X, Kang L, Luo X, Zhu W, Chen K, Wang X, Jiang H. PDTD: a web-accessible protein database for drug target identification. BMC Bioinformatics. 2008;9:104. doi: 10.1186/1471-2105-9-104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Li H, Gao Z, Kang L, Zhang H, Yang K, Yu K, Luo X, Zhu W, Chen K, Shen J, et al. TarFisDock: a web server for identifying drug targets with docking approach. Nucleic Acids Res. 2006;34:W219–W224. doi: 10.1093/nar/gkl114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kuhn M, Szklarczyk D, Franceschini A, Campillos M, von Mering C, Jensen LJ, Beyer A, Bork P. STITCH 2: an interaction network database for small molecules and proteins. Nucleic Acids Res. 2010;38:D552–D556. doi: 10.1093/nar/gkp937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gunther S, Kuhn M, Dunkel M, Campillos M, Senger C, Petsalaki E, Ahmed J, Urdiales EG, Gewiess A, Jensen LJ, et al. SuperTarget and Matador: resources for exploring drug-target relationships. Nucleic Acids Res. 2008;36:D919–D922. doi: 10.1093/nar/gkm862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lim E, Pon A, Djoumbou Y, Knox C, Shrivastava S, Guo AC, Neveu V, Wishart DS. T3DB: a comprehensively annotated database of common toxins and their targets. Nucleic Acids Res. 2010;38:D781–D786. doi: 10.1093/nar/gkp934. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Sperandio O, Petitjean M, Tuffery P. wwLigCSRre: a 3D ligand-based server for hit identification and optimization. Nucleic Acids Res. 2009;37:W504–W509. doi: 10.1093/nar/gkp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Toomey D, Hoppe HC, Brennan MP, Nolan KB, Chubb AJ. Genomes2Drugs: identifies target proteins and lead drugs from proteome data. PLoS One. 2009;4:e6195. doi: 10.1371/journal.pone.0006195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Strohl WR. The role of natural products in a modern drug discovery program. Drug Discov. Today. 2000;5:39–41. doi: 10.1016/s1359-6446(99)01443-9. [DOI] [PubMed] [Google Scholar]
- 11.Ehrman TM, Barlow DJ, Hylands PJ. Phytochemical databases of Chinese herbal constituents and bioactive plant compounds with known target specificities. J. Chem. Inf. Model. 2007;47:254–263. doi: 10.1021/ci600288m. [DOI] [PubMed] [Google Scholar]
- 12.Buckingham J. Dictionary of Natural Products. London: Chapman and Hall; 1994. [Google Scholar]
- 13.Wang JF, Zhou H, Han LY, Chen X, Chen YZ, Cao ZW. Traditional Chinese medicine information database. Clin. Pharmacol. Ther. 2005;78:92–93. doi: 10.1016/j.clpt.2005.03.010. [DOI] [PubMed] [Google Scholar]
- 14.Zhou J, Yan X, Xie G. Traditional Chinese Medicines: Molecular Structures, Natural Sources & Applications. 2nd edn. 2003. Ashgate/Aldershot/Hampshire/ England/Burlington, VT, USA. [Google Scholar]
- 15.Kanehisa M, Araki M, Goto S, Hattori M, Hirakawa M, Itoh M, Katayama T, Kawashima S, Okuda S, Tokimatsu T, et al. KEGG for linking genomes to life and the environment. Nucleic Acids Res. 2008;36:D480–D484. doi: 10.1093/nar/gkm882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Willett P, Barnard JM, Downs GM. Chemical similarity searching. J. Chem. Inform. Comput. Sci. 1998;38:983–996. [Google Scholar]
- 17.Bostrom J, Hogner A, Schmitt S. Do structurally similar ligands bind in a similar fashion? J. Med. Chem. 2006;49:6716–6725. doi: 10.1021/jm060167o. [DOI] [PubMed] [Google Scholar]
- 18.Huang N, Shoichet BK, Irwin JJ. Benchmarking sets for molecular docking. J. Med. Chem. 2006;49:6789–6801. doi: 10.1021/jm0608356. [DOI] [PMC free article] [PubMed] [Google Scholar]