Abstract
The fungal genus Fusarium includes many plant and/or animal pathogenic species and produces diverse toxins. Although accurate species identification is critical for managing such threats, it is difficult to identify Fusarium morphologically. Fortunately, extensive molecular phylogenetic studies, founded on well-preserved culture collections, have established a robust foundation for Fusarium classification. Genomes of four Fusarium species have been published with more being currently sequenced. The Cyber infrastructure for Fusarium (CiF; http://www.fusariumdb.org/) was built to support archiving and utilization of rapidly increasing data and knowledge and consists of Fusarium-ID, Fusarium Comparative Genomics Platform (FCGP) and Fusarium Community Platform (FCP). The Fusarium-ID archives phylogenetic marker sequences from most known species along with information associated with characterized isolates and supports strain identification and phylogenetic analyses. The FCGP currently archives five genomes from four species. Besides supporting genome browsing and analysis, the FCGP presents computed characteristics of multiple gene families and functional groups. The Cart/Favorite function allows users to collect sequences from Fusarium-ID and the FCGP and analyze them later using multiple tools without requiring repeated copying-and-pasting of sequences. The FCP is designed to serve as an online community forum for sharing and preserving accumulated experience and knowledge to support future research and education.
INTRODUCTION
The fungal genus Fusarium poses a multifaceted threat to global crop production and animal/human health. Collectively, the genus includes many important plant pathogens (1). Certain Fusarium secondary metabolites, such as fumonisins, trichothecenes, enniatins and zearalenone, are toxins that threaten food safety and animal/human health (2). Some species infect immune-compromised individuals (3,4) but also cause corneal infections in people with healthy immune systems (5,6). Due to its practical importance, the genus has been extensively studied at levels ranging from genetic mechanisms underlying important traits, such as toxin production and pathogenicity, to global biodiversity and evolution (2,7–11).
More than 35 000 strains isolated from various substrates around the world are accessioned in the Fusarium Research Center (FRC) and the USDA-ARS National Center for Agricultural Utilization Research (NCAUR) Culture Collection, making this genus the best-preserved fungal group. Using this rich strain resource, extensive molecular phylogenetic studies have been conducted, resulting in data covering most agriculturally and/or medically important species complexes (6,9,10,12–22). However, despite these advances, a significant amount of diversity has yet to be explored, and some species complexes are quite poorly characterized phylogenetically. To support and coordinate the remaining phylogenetic analyses, it is essential to archive available phylogenetic data and associated cultures in a format that is readily accessible and searchable by members of the global Fusarium research community.
In 2004, we released Fusarium-ID, a simple, web-accessible BLAST server that consisted of sequences of the translation elongation factor 1α (EF-1α) gene from a fairly representative spectrum of Fusarium species (13). Since then, we have expanded Fusarium-ID to include sequences of multiple marker loci that represent almost all known species and to provide more data analysis and visualization tools. We also developed two additional platforms, including the Fusarium Comparative Genomics Platform (FCGP) and the Fusarium Community Platform (FCP), to build a more comprehensive community resource, named as the Cyber infrastructure for Fusarium (CiF; http://www.fusariumdb.org/; Figure 1). The main motivation for building CiF was to support the archiving and integration of data and knowledge from disparate yet related areas of research on Fusarium through a single-integrated platform. Many informatics platforms supporting fungal research have been developed. However, because they are often specialized for only a subset of data (e.g. genome sequences, data associated with culture collections or specific gene families), integrated analysis of disparate data sets across multiple taxa is cumbersome, as data from multiple sources need to be mined and integrated in an ad hoc manner. With diverse, systematically archived data sets, the CiF aims to efficiently leverage new knowledge and support problem solving. We have been building platforms similar to CiF to support research and education on other taxa [e.g. the Phytophthora Database (23)]. These platforms share common architecture and tools, such that building new platforms and improving existing platforms is efficient and cost-effective. As more platforms are added, they will form a comprehensive cyber infrastructure supporting fungal research.
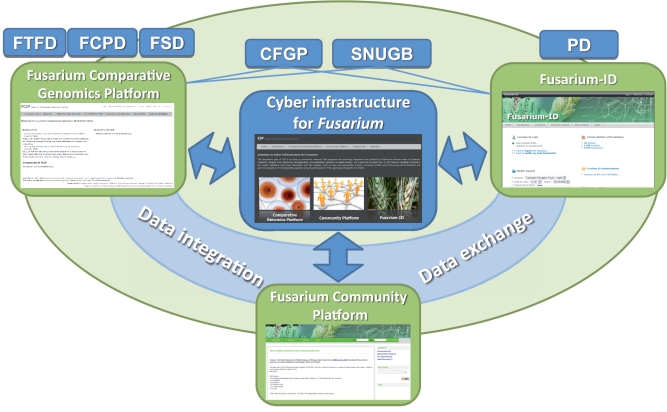
Figure 1.
System structure of the CiF. The CiF consists of three components, FCGP, Fusarium-ID and FCP. Several databases specialized for fungal genome sequences or gene families and functional groups, including Fungal Transcription Factor Database (FTFD), Fungal Cytochrome P450 Database (FCPD), Fungal Secretome Database (FSD) and Comparative Fungal Genomics Platform (CFGP), feed data to the FCGP. Data analysis and visualization tools from the CFGP, the Seoul National University Genome Browser (SNUGB) and Phytophthora Database (PD) support data visualization and analysis.
FUSARIUM-ID
With the advent of molecular tools and robust molecular evolutionary principles, it has become easier and faster to recognize new species when they are encountered (24). Systematically archiving available Fusarium phylogenetic data will help guide future species descriptions, coordinate community research on its systematics, and support education in Fusarium biology. Without a robust phylogenetic framework and community-wide knowledge sharing, discovery and characterization of novel Fusarium species will likely be fragmented, creating confusion instead of the order that taxonomy should provide. The Fusarium-ID (http://isolate.fusariumdb.org/) consists of a database of extensive sequence data from most known Fusarium species and data analysis and visualization tools. The Fusarium-ID enables users to explore the diversity of Fusarium and accurately identify new isolates based on their sequence similarity to previously characterized species.
Data content and utility
The Fusarium-ID currently archives 5558 marker sequences from 1844 isolates representing over 200 phylogenetically distinct species. Its data content will grow rapidly, as we continuously curate and deposit data from previous and current phylogenetic studies. All sequence data in this database have been derived from vouchered, publicly available cultures, allowing users to further investigate any connections between their query and hits in the database. Most of the data in the Fusarium-ID database are also available and searchable through the Centraalbureau voor Schimmelcultures (CBS) Fungal Biodiversity Center (http://www.cbs.knaw.nl/fusarium/) so that they will be maintained in multiple electronic resources around the world.
Sequence data from more than 10 marker loci are archived in the Fusarium-ID database with some loci sequenced at multiple locations (e.g. ribosomal RNA encoding genes and their spacer regions), but individual species typically have been characterized only by a subset of these markers. However, sequences at three loci, including EF-1α and two genes encoding the largest and second largest subunits of RNA polymerase (RPB1 and RPB2, respectively), are being generated for all of the phylogenetic species, and can serve as markers individually and collectively to help identify new isolates to the species level. To identify a new isolate, sequence data from one or more of these genes can be used as a BLAST query against the Fusarium-ID sequence database. Considering the extensive coverage of human pathogenic species in the database, an exact match can be reasonably interpreted as definite species-level identification (10). However, because the value of a sequence match result depends on a few experimental or biological factors, all results must be interpreted with care (10,24). Precise conclusions may require phylogenetic analysis based on multiple markers, especially when the BLAST results suggest that the query sequence may represent a novel species not currently represented in Fusarium-ID. In such cases, users should employ an appropriate multilocus typing scheme to assess genealogical concordance and evidence of genetic exchange (25,26). Individual sequences and sequence alignments from previous phylogenetic analyses can be downloaded from Fusarium-ID so that users can conduct their own analyses, and appropriate cultures can be ordered from FRC and/or the USDA-ARS NCAUR Culture Collection.
Data analysis and visualization tools
In addition to BLAST, Fusarium-ID provides a number of functions adopted from the Phytophthora Database (http://www.phytophthoradb.org/), a platform we developed to support the identification and monitoring of Phytophthora species and populations (23). Via the Folder function, users can create two types of data storage space in the CiF: (i) private folder for storing selected data and results from previous analyses and (ii) shared folder that permits data sharing with other users designated by the creator of the folder (by assigning user IDs permitted to access the folder). The shared folder function enables communication and collaboration among multiple users via the CiF. The Cart function described below allows users to collect data in multiple areas of Fusarium-ID and use/analyze them later. A suite of web tools, named the Phyloviewer, allows users to align sequences in BLAST outputs, including the query sequence, and any data stored in the Cart and build phylogenetic trees on the fly. Sequence data in the resulting tree are linked to information associated with corresponding isolates so that users can browse if any notable patterns (e.g. geographic and host of origins, mycotoxin profiles, etc) exist among the isolates included in the tree. The Virtual Gel function supports this diagnostic method by generating predicted restriction fragment length polymorphism (RFLP) patterns from chosen sequences and restriction enzyme(s) via a virtual gel.
Future plans
The CiF data warehouse will be continuously populated with phylogenetic sequence data from our previous and current studies to provide a robust foundation for ecological and phylogenetic studies and genome sequencing efforts. The utility of Fusarium-ID will be enhanced as members of the global Fusarium research community deposit cultures of novel species, along with associated sequence chromatograms and data, so that the sequence results can be verified and isolates are made available for future study. We also plan to add photographs and/or line drawings illustrating key morphological features associated with each phylogenetic species.
Two main functions that will be available soon are a geographic information system (GIS) tool and a tool for searching and comparing population genetic diversity data based on simple sequence repeat (SSR) loci. Both tools are currently functional in the Phytophthora Database. The GIS tool will function as a digitized atlas showing the genotypic and phenotypic diversity of Fusarium worldwide in geospatial and temporal contexts. This functionality will help to establish a baseline for monitoring the biogeographic diversity of Fusarium species. For major pathogenic Fusarium species, we plan to generate MLST (Multi-Locus Sequence Typing) data sets for population-level analyses. In combination with the GIS tool and a search tool for MLST data (to be developed), users can significantly increase their sampling by integrating their datasets with those available in Fusarium-ID, monitor haplotype diversity across hosts and geographic regions, and examine the demographic history of species/populations.
FCGP
Rapidly accumulating genome sequence data from diverse Fusarium species with different traits offers tremendous opportunities for understanding the molecular and evolutionary mechanisms underpinning functional diversification at a genome level (8,27,28). The FCGP (http://genomics.fusariumdb.org/) was developed to facilitate the realization of such opportunities. Besides providing an interactive genome browser, the FCGP presents computed characteristics of multiple gene families and functional groups in sequenced species to support quick comparison and analysis across species. In combination with the phylogenetic framework and accessioned cultures available through Fusarium-ID, the FCGP will help users study the evolution of Fusarium genes, gene networks and whole genomes.
Genome data
The genomes of four Fusarium species, including F. graminearum (two strains), F. oxysporum, F. verticillioides and one species in the F. solani species complex, have been sequenced (8,27,28) with more species and isolates currently being sequenced or annotated. The first three species were sequenced by the Broad Institute (http://www.broadinstitute.org/annotation/genome/fusarium_group/MultiHome.html), while the Department of Energy Joint Genome Institute sequenced F. solani (also known as Nectria haematococca Mating Population VI; http://genome.jgi-psf.org/Necha2/Necha2.home.html). We converted sequences from these sites into a common format via the data extraction pipeline of the Comparative Fungal Genomics Platform (CFGP; http://cfgp.snu.ac.kr/) (29). The most recent versions of genome data, as well as earlier versions, are available in the FCGP (under the ‘List of all Fusarium genomes’ in the ‘GENOMEs’ menu). Results of the annotation of each gene with 12 different programs, including InterPro scans, subcelluar localization prediction softwares, signal-peptide prediction programs and transmembrane helix prediction programs, are presented.
Genome browser
As the scale and complexity of genome sequence analyses increases, a versatile genome browser has become an essential tool. To support visualization and utilization of genome sequences and features both within and across species, the SNU Genome Browser (SNUGB; http://genomebrowser.snu.ac.kr/) (30), a genome browser developed to support the CFGP and several platforms derived from the CFGP, was integrated into the FCGP. The application interface of the SNUGB was designed in a modular fashion to facilitate the addition of new tools and its customization for specialized platforms. The SNUGB has already been implemented in multiple platforms (31–37). All sequence data and contig information are displayed through the interface of the Contig Browser. Annotation information in a chosen region, such as transcripts, ORFs, tRNAs/rRNAs, exon/intron structure, SignalP, PSort and InterPro domains, can be displayed in multiple formats. Such information will be useful for in-depth analysis of gene function. In addition, the Chromosome Viewer shows the chromosomal locations of the phylogenetic markers stored in Fusarium-ID. Once the user clicks the bar indicating a specific marker, genomic sequences and features around the marker can be viewed through the SNUGB interface. The SNUGB can display genome sequences from multiple strains/species around a chosen locus to facilitate the evaluation and development of new phylogenetic markers, including the design of PCR primers.
Platforms that support the analysis of specific gene families and functional groups
We have constructed several comparative genomics platforms specialized for supporting in-depth analysis of specific gene families and functional groups in fungi (31,35,36). One of them is the Fungal Transcription Factor Database (FTFD; http://ftfd.snu.ac.kr/) (36), in which all putative transcription factors (TFs) encoded by sequenced fungal and oomycete species were identified and classified into families. Cytochrome P450s, a superfamily of heme-containing monooxygenases, play critical roles in fungal metabolism and ecology by participating in the production of diverse metabolites and also modifying harmful environmental chemicals (38). The Fungal Cytochrome P450 Database (FCPD; http://p450.riceblast.snu.ac.kr/) (35) archives genes encoding P450s to support studies on their function and evolution. The Fungal Secretome Database (FSD; http://fsd.snu.ac.kr/) identifies and archives putative secretory proteins (31). Fusarium-specific data from these three platforms, including 3075 TFs, 577 cytochrome P450s and 11668 putative secretory proteins, are organized to provide an overview of these proteins within and across Fusarium species. A BLAST server for each data set is available for quick search. Moreover, genes that appear unique to each species, as well as those that are present in subsets of the four species, were identified through BLASTMatrix2, a modified BLAST program that searches gene(s) homologous to a query in multiple species simultaneously. With FTFD, FCPD, FSD and BLASTMatrix2, Fusarium proteins can be quickly compared with those in other fungal taxa.
Future plans
In addition to depositing newly released Fusarium genome sequences, characteristics of additional protein groups, such as ABC transporters and carbohydrate degrading enzymes, will be added once the corresponding fungal kingdom-wide databases are established. Available expressed sequence tags from Fusarium species will also be archived and linked to the corresponding genomes.
CART/FAVORITE: A HUB FOR DATA REPOSITORY AND ANALYSIS
Availability of multiple disparate, yet complementary, data through a single platform opens up the possibility of integrated analysis without going through data retrieval from multiple independent sources. However, to enable such analysis, a tool for retrieving and managing data from multiple databases is required. The Cart (or ‘Favorite’ in the FCGP) function serves as such a tool (Figure 2). Through this function, users can collect metadata for any sequences deposited in Fusarium-ID and the FCPD, and all or part of the collected data can be analyzed later by available data analysis tools, including BLAST, ClustalW, Virtual Gel, and Primer3, without requiring repeated copying-and-pasting of sequences for different analyses. Each cart/favorite can be stored in user's private and/or shared folder. The Cart (Favorite) function has been implemented in several platforms we have developed (29,31,35,36). Many additional analysis tools that are currently operational in the CFGP (29) will be integrated to the CiF Cart/Favorite function to expand its utility.
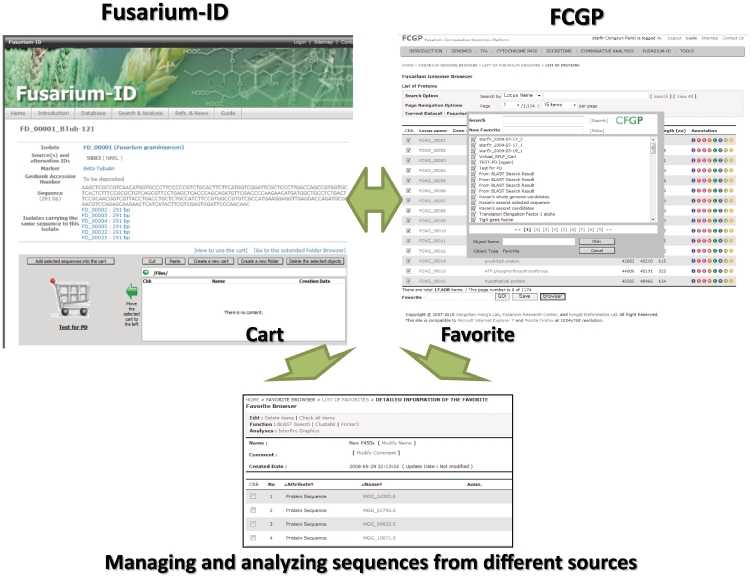
Figure 2.
Cart/Favorite, a hub for data storage and analysis in the CiF. Users can collect sequences through two different interfaces: (i) Cart in Fusarium-ID and (ii) Favorite in the FCGP. All or part of the sequences collected in a cart/favorite, can be analyzed using several bioinformatics tools and can also be shared with other users through the Folder function.
FCP
Sharing experience and knowledge is fundamental to help leverage new knowledge and educate the next generation of researchers and educators. Without an efficient mechanism to support such effort, scientific endeavors will be fragmented and become inefficient. Web 2.0 technologies offer immense potential for supporting the pooling and sharing of diverse experience and knowledge accumulated in a global research community without being limited by distance and traditional forms of organizational structure. A number of scientific communities have built a web platform and associated databases and material resources to support community research and education. Some notable examples include the Arabidopsis Information Resource (TAIR; http://www.arabidopsis.org/), the iPlant Collaborative (iPlant; http://www.iplantcollaborative.org/), the PHI-base (http://www.phi-base.org/about.php), GMOD (http://gmod.org/wiki/Main_Page) and the Aspergillus website (http://www.aspergillus.org.uk/). However, this potential needs to be more widely harnessed.
The FCP (http://blog.fusariumdb.org/) is currently being developed to support the preservation and sharing of experience and knowledge accumulated in the global Fusarium research community. Because the FCP is a new addition to the CiF, the currently available content is rather limited. However, through ‘crowd sourcing’ in the community, we plan to expand its content quickly. The FCP is expected to provide quick reviews of latest research development, experimental protocols, educational modules, and community news via a blog interface. The trail of communications associated with the archived content, particularly protocols, will help new comers to the community quickly learn from the collective knowledge rather than learning them through trial and error. Because it is web based, the FCP will also become an ideal medium for rapidly sharing information on emerging disease problems and coordinating subsequent responses. We envision that the FCP will also function to support global human networking.
AVAILABILITY AND REQUIREMENTS
All three platforms of the CiF can be accessed via the gateway web page (http://www.fusariumdb.org/). Although registration is not mandatory, certain functions, such as Folder and Cart and writing a post through the FCP, are not available without login. More than 250 users from 33 countries have registered. However, considering that guests can access most functions and data, the actual number of users and countries should be larger. User access data (monthly and yearly) can be found in the statistics page of the CiF.
FUNDING
This work was indirectly supported by grants from the United States Department of Agriculture-AFRI Plant Biosecurity program (grant numbers 2005-35605-15393 and 2008-55605-18773). These grants enabled the development of the Phytophthora Database. Tools and experience from the Phytophthora Database greatly facilitated the construction of the CiF. This work also was partially supported by the following grants to Y.-H.L.; National Research Foundation of Korea (grant numbers 2009-0063340 and 2009-0080161); Biogreen21 (grant number 20080401-034-044-009-01-00); Technology Development Program for Agriculture and Forestry (grant number 309015-04-SB020); Crop Functional Genomics Center (grant number 2009K001198). Funding for open access charge: Penn State University.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Stacy Sink and Jean H. Juba for excellent technical assistance, Nathane Orwig for generating the DNA sequences in National Center for Agricultural Utilization Research (NCAUR)'s DNA core facility and individuals who have deposited Fusarium isolates to FRC and/or USDA-ARS-NCAUR.
REFERENCES
- 1.Agrios GN. Plant Pathology. 5 edn. San Diego: Academic Press; 2005. [Google Scholar]
- 2.Desjardins AE. Fusarium Mycotoxins: Chemistry, Genetics & Biology. St. Paul, MN: APS Press; 2006. [Google Scholar]
- 3.Anaissie E, Kantarjian H, Ro J, Hopfer R, Rolston K, Fainstein V, Bodey G. The emerging role of Fusarium infections in patients with cancer. Medicine. 1988;67:77–83. doi: 10.1097/00005792-198803000-00001. [DOI] [PubMed] [Google Scholar]
- 4.Dignani MC, Anaissie E. Human fusariosis. Clin. Microbiol. Infect. 2004;10(Suppl. 1):67–75. doi: 10.1111/j.1470-9465.2004.00845.x. [DOI] [PubMed] [Google Scholar]
- 5.Chang DC, Grant GB, O'Donnell K, Wannemuehler KA, Noble-Wang J, Rao CY, Jacobson LM, Crowell CS, Sneed RS, Lewis FMT, et al. Multistate outbreak of Fusarium keratitis associated with use of a contact lens solution. JAMA. 2006;296:953–963. doi: 10.1001/jama.296.8.953. [DOI] [PubMed] [Google Scholar]
- 6.O'Donnell K, Sarver B, Brandt M, Chang DC, Nobel-Wang J, Park BJ, Sutton D, Benjamin L, Lindsley M, Padhye A, et al. Phylogenetic diversity and microsphere array-based genotyping of human pathogenic fusaria, including isolates from the multistate contact lens-associated U.S. keratitis outbreaks of 2005 and 2006. J. Clin. Microbiol. 2007;45:2235–2248. doi: 10.1128/JCM.00533-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Balmas V, Migheli Q, Scherm B, Garau P, O'Donnell K, Ceccherelli G, Kang S, Geiser DM. Multilocus phylogenetics show high levels of endemic fusaria inhabiting Sardinian soils (Tyrrhenian Islands) Mycologia. 2010;102:803–812. doi: 10.3852/09-201. [DOI] [PubMed] [Google Scholar]
- 8.Ma L-J, van der Does HC, Borkovich KA, Coleman JJ, Daboussi M-J, Di Pietro A, Dufresne M, Freitag M, Grabherr M, Henrissat B, et al. Comparative genomics reveals mobile pathogenicity chromosomes in Fusarium. Nature. 2010;464:367–373. doi: 10.1038/nature08850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.O'Donnell K, Ward TJ, Geiser DM, Kistler HC, Aoki T. Genealogical concordance between the mating type locus and seven other nuclear genes supports formal recognition of nine phylogenetically distinct species within the Fusarium graminearum clade. Fungal Genet. Biol. 2004;41:600–623. doi: 10.1016/j.fgb.2004.03.003. [DOI] [PubMed] [Google Scholar]
- 10.O'Donnell K, Sutton DA, Rinaldi MG, Sarver BAJ, Balajee SA, Schroers H-J, Summerbell RC, Robert VARG, Crous PW, Zhang N, et al. An internet-accessible DNA sequence database for identifying fusaria from human and animal infections. J. Clin. Microbiol. 2010;48:3708–3718. doi: 10.1128/JCM.00989-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Summerell BA, Leslie JF, Backhouse D, Bryden WL, Burgess LW, editors. Fusarium: Paul E. Nelson Memorial Symposium. St. Paul, MN: APS Press; 2001. [Google Scholar]
- 12.Aoki T, O'Donnell K, Scandiani MM. Sudden death syndrome of soybean in South America is caused by four species of Fusarium: Fusarium brasiliense sp. nov., F. cuneirostrum sp. nov., F. tucumaniae and F. virguliforme. Mycoscience. 2005;46:162–183. [Google Scholar]
- 13.Geiser DM, Jiménez-Gasco M, Kang S, Makalowska I, Veeraraghavan N, Ward TJ, Zhang N, Kuldau GA, O'Donnell K. FUSARIUM-ID v.1.0: a DNA sequence database for identifying Fusarium. Eur. J. Plant Pathol. 2004;110:473–479. [Google Scholar]
- 14.O'Donnell K, Cigelnik E, Nirenberg HI. Molecular systematics and phylogeography of the Gibberella fujikuroi species complex. Mycologia. 1998;90:465–493. [Google Scholar]
- 15.O'Donnell K, Kistler HC, Cigelnik E, Ploetz RC. Multiple evolutionary origins of the fungus causing Panama disease of banana: concordant evidence from nuclear and mitochondrial gene genealogies. Proc. Natl Acad. Sci. USA. 1998;95:2044–2049. doi: 10.1073/pnas.95.5.2044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.O'Donnell K, Kistler HC, Tacke BK, Casper HH. Gene genealogies reveal global phylogeographic structure and reproductive isolation among lineages of Fusarium graminearum, the fungus causing wheat scab. Proc. Natl Acad. Sci. USA. 2000;97:7905–7910. doi: 10.1073/pnas.130193297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.O'Donnell K, Nirenberg HI, Aoki T, Cigelnik E. A multigene phylogeny of the Gibberella fujikuroi species complex: detection of additional phylogenetically distinct species. Mycoscience. 2000;41:61–78. [Google Scholar]
- 18.O'Donnell K, Sutton DA, Rinaldi MG, Magnon KC, Cox PA, Revankar SG, Sanche S, Geiser DM, Juba JH, van Burik J-AH, et al. Genetic diversity of human pathogenic members of the Fusarium oxysporum complex inferred from multilocus DNA sequence data and amplified fragment length polymorphism analyses: evidence for the recent dispersion of a geographically widespread clonal lineage and nosocomial origin. J. Clin. Microbiol. 2004;42:5109–5120. doi: 10.1128/JCM.42.11.5109-5120.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Starkey DE, Ward TJ, Aoki T, Gale LR, Kistler HC, Geiser DM, Suga H, Tóth B, Varga J, O'Donnell K. Global molecular surveillance reveals novel Fusarium head blight species and trichothecene toxin diversity. Fungal Genet. Biol. 2007;44:1191–1204. doi: 10.1016/j.fgb.2007.03.001. [DOI] [PubMed] [Google Scholar]
- 20.Zhang N, O'Donnell K, Sutton DA, Nalim FA, Summerbell RC, Padhye AA, Geiser DM. Members of the Fusarium solani species complex that cause infections in both humans and plants are common in the environment. J. Clin. Microbiol. 2006;44:2186–2190. doi: 10.1128/JCM.00120-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schroers H-J, O'Donnell K, Lamprecht SC, Kammeyer PL, Johnson S, Sutton DA, Rinaldi MG, Geiser DM, Summerbell RC. Taxonomy and phylogeny of the Fusarium dimerum species group. Mycologia. 2009;101:44–70. doi: 10.3852/08-002. [DOI] [PubMed] [Google Scholar]
- 22.O'Donnell K, Gueidan C, Sink S, Johnston PR, Crous PW, Glenn A, Riley R, Zitomer NC, Colyer P, Waalwijk C, et al. A two-locus DNA sequence database for typing plant and human pathogens within the Fusarium oxysporum species complex. Fungal Genet. Biol. 2009;46:936–948. doi: 10.1016/j.fgb.2009.08.006. [DOI] [PubMed] [Google Scholar]
- 23.Park J, Park B, Veeraraghavan N, Jung K, Lee Y-H, Blair JE, Geiser DM, Isard S, Mansfield MA, Nikolaeva E, et al. Phytophthora database: a forensic database supporting the identification and monitoring of Phytophthora. Plant Disease. 2008;92:966–972. doi: 10.1094/PDIS-92-6-0966. [DOI] [PubMed] [Google Scholar]
- 24.Kang S, Mansfield MA, Park B, Geiser DM, Ivors KL, Coffey MD, Grunwald N, Martin FM, Levesque A, Blair JE. The promise and pitfalls of sequence-based identification of plant pathogenic fungi and oomycetes. Phytopathology. 2010;100:732–737. doi: 10.1094/PHYTO-100-8-0732. [DOI] [PubMed] [Google Scholar]
- 25.Geiser DM. In: Advances in Fungal Biotechnology for Industry. Tkacz J, Lange L, editors. Dordrecht: Kluwer Academic Publishers; 2004. pp. 1–12. [Google Scholar]
- 26.Taylor JW, Jacobson DJ, Kroken S, Kasuga T, Geiser DM, Hibbett DS, Fisher MC. Phylogenetic species recognition and species concepts in fungi. Fungal Genet. Biol. 2000;31:21–32. doi: 10.1006/fgbi.2000.1228. [DOI] [PubMed] [Google Scholar]
- 27.Coleman JJ, Rounsley SD, Rodriguez-Carres M, Kuo A, Wasmann CC, Grimwood J, Schmutz J, Taga M, White GJ, Zhou S, et al. The genome of Nectria haematococca: contribution of supernumerary chromosomes to gene expansion. PLoS Genet. 2009;5:e1000618. doi: 10.1371/journal.pgen.1000618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Cuomo CA, Guldener U, Xu J-R, Trail F, Turgeon BG, Di Pietro A, Walton JD, Ma L-J, Baker SE, Rep M, et al. The Fusarium graminearum Genome Reveals a Link Between Localized Polymorphism and Pathogen Specialization. Science. 2007;317:1400–1402. doi: 10.1126/science.1143708. [DOI] [PubMed] [Google Scholar]
- 29.Park J, Park B, Jung K, Jang S, Yu K, Choi J, Kong S, Park J, Kim S, Kim H, et al. CFGP: a web-based, comparative fungal genomics platform. Nucl. Acids Res. 2008;36:D562–571. doi: 10.1093/nar/gkm758. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jung K, Park J, Choi J, Park B, Ahn K, Choi J, Kim S, Choi D, Kang S, Lee Y-H. SNUGB: a versatile genome browser supporting comparative and functional fungal genomics. BMC Bioinformatics. 2008;9:586. doi: 10.1186/1471-2164-9-586. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Choi J, Park J, Kim D, Jung K, Kang S, Lee Y-H. Fungal Secretome Database: integrated platform for annotation of fungal secretomes. BMC Genomics. 2010;11:105. doi: 10.1186/1471-2164-11-105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Choi J, Park J, Jeon J, Chi MH, Goh J, Yoo SY, Park J, Jung K, Kim H, Park SY, et al. Genome-wide analysis of T-DNA integration into the chromosomes of Magnaporthe oryzae. Mol. Microbiol. 2007;66:371–382. doi: 10.1111/j.1365-2958.2007.05918.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jeon J, Park SY, Chi MH, Choi J, Park J, Rho HS, Kim S, Goh J, Yoo S, Choi J, et al. Genome-wide functional analysis of pathogenicity genes in the rice blast fungus. Nat. Genet. 2007;39:561–565. doi: 10.1038/ng2002. [DOI] [PubMed] [Google Scholar]
- 34.Lee W, Park J, Choi J, Jung K, Park B, Kim D, Lee J, Ahn K, Song W, Kang S, et al. IMGD: an integrated platform supporting comparative genomics and phylogenetics of insect mitochondrial genomes. BMC Genomics. 2009;10:148. doi: 10.1186/1471-2164-10-148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Park J, Choi J, Ahn K, Park B, Park J, Kang S, Lee Y-H. Fungal cytochrome P450 database. BMC Bioinformatics. 2008;9:402. doi: 10.1186/1471-2164-9-402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park J, Park J, Jang S, Kim S, Kong S, Choi J, Ahn K, Kim J, Lee S, Kim S, et al. FTFD: an informatics pipeline supporting phylogenomic analysis of fungal transcription factors. Bioinformatics. 2008;24:1024–1025. doi: 10.1093/bioinformatics/btn058. [DOI] [PubMed] [Google Scholar]
- 37.Xi H, Park J, Ding G, Lee Y-H, Li Y. SysPIMP: the web-based systematical platform for identifying human disease-related mutated sequences from mass spectrometry. Nucl. Acids Res. 2009;37:D913–D920. doi: 10.1093/nar/gkn848. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.van den Brink HM, van Gorcom RFM, van den Hondel CAMJJ, Punt PJ. Cytochrome P450 enzyme systems in fungi. Fungal Genet. Biol. 1998;23:1–17. doi: 10.1006/fgbi.1997.1021. [DOI] [PubMed] [Google Scholar]