Abstract
By broad literature survey, we have developed a leaf senescence database (LSD, http://www.eplantsenescence.org/) that contains a total of 1145 senescence associated genes (SAGs) from 21 species. These SAGs were retrieved based on genetic, genomic, proteomic, physiological or other experimental evidence, and were classified into different categories according to their functions in leaf senescence or morphological phenotypes when mutated. We made extensive annotations for these SAGs by both manual and computational approaches, and users can either browse or search the database to obtain information including literatures, mutants, phenotypes, expression profiles, miRNA interactions, orthologs in other plants and cross links to other databases. We have also integrated a bioinformatics analysis platform WebLab into LSD, which allows users to perform extensive sequence analysis of their interested SAGs. The SAG sequences in LSD can also be downloaded readily for bulk analysis. We believe that the LSD contains the largest number of SAGs to date and represents the most comprehensive and informative plant senescence-related database, which would facilitate the systems biology research and comparative studies on plant aging.
INTRODUCTION
Leaf senescence is the last phase of plant development and a highly coordinated process regulated by a large number of senescence associated genes (SAGs) (1–2). Leaf senescence can either be naturally induced during development stages, or stimulated by environmental factors including darkness, nutritional deficiency and various stresses (1–3). Premature senescence is an important factor leading to the decrease of crop yield and quality, which becomes an increasing concern due to the global climate change in the recent years.
Many advances in the understanding of leaf senescence at the molecular level had been achieved through the identification and characterization of hundreds of SAGs and senescence-related mutants in Arabidopsis thaliana, Lycopersicon esculentum and Nicotiana tabaccum (1–4). Microarray expression profiling in Arabidopsis revealed that more than 800 genes are up-regulated during the course of leaf senescence (5). Among them, more than 200 transcription factors, including WRKY, NAC, MADS, MYB, bZIP and bHLH family members, are involved in the regulation of leaf senescence, indicating that leaf senescence is governed by complex transcriptional regulatory networks.
Molecular and genetic studies of leaf senescence in recent years led to the accumulation of a large volume of scattered information related to SAGs. The construction of a leaf senescence related database with wide-spread collection and systematic annotation of SAGs may provide a useful resource and a good starting point for the further study of the molecular aspects of leaf senescence. Some initial efforts to this end had been made; including the online website of Plant Senescence Network (SenNet) constructed by Thomas and his colleagues (http://www.sidthomas.net/Plant_senescence/) and the corresponding SenWiki web pages (http://www.sidthomas.net/SenWiki/). SenNet brings together various community information including meetings, laboratories, websites and useful links related to plant senescence, while SenWiki provides general information and knowledge of senescence with texts and images. However, a wide-ranging compilation and detailed annotation of known SAGs that would be of great help and demand to the systemic study of leaf senescence at the molecular level has yet to be done. In this regard, we have developed a leaf senescence database (LSD) (http://www.eplantsenescence.org/) by retrieving and integrating information from research papers and public databases.
At present 1145 SAGs from 21 species were manually curated and categorized into several groups according to their function. Users can browse the entries in the database to obtain information including literatures, mutants, phenotypes, expression profiles, miRNA interactions, orthologs in other plants and cross links to protein domain and family databases. Users can also search the database easily through the ‘Text Search’ interface with locus names, keywords and author names, etc. We have implemented the BLAST tool kit for sequence similarity search against nucleotide or protein sequences of these SAGs in the LSD. A major feature of the LSD is the integration of a bioinformatics platform WebLab we had developed previously (6), which allows users to perform extensive sequence analysis of the SAGs they are interested in. All the SAG sequences are freely available for downloading. Help information including user guides, a tutorials and FAQs are also available online. We plan to identify putative SAGs from completely sequenced genomes and add them into the database in the near future, and improve the user interface based on user feedback, provide more help documents with case studies, add more links to other useful sites, and update the database in time with more leaf senescence related data available. We hope that LSD could be a useful resource for the leaf senescence research community, as well as a gateway for the collaborative project we are working with both domestic and international colleagues.
IDENTIFICATION AND ANNOTATION OF SAGS
We made an extensive literature survey and collected approximately 200 leaf senescence related papers published before October 2010 on major plant biology journals including Plant Cell, Plant Physiology, Plant Journal, New Phytologist, Journal of Experimental Botany, among others. A total of 1145 SAGs from 21 species including Arabidopsis, rice and so on, were identified and manually verified based on genetic, genomic, proteomic, physiological and other experimental evidence. Among them, 96 and 58 genes were supported by mutational investigation or transgenic over-expression study, respectively (Table 1).
Table 1.
Number of SAGs and mutants in 21 species
| Species | Common name | SAGs | Mutants | Transgenic |
|---|---|---|---|---|
| Arabidopsis thaliana | Thale Cress | 949 | 90 | 35 |
| Oryza sativa | Rice | 104 | 3 | 9 |
| Medicago truncatula | Barrel Clover | 31 | 0 | 0 |
| Brassica napus | Rape | 15 | 0 | 0 |
| Lycopersicon esculentum | Tomato | 8 | 0 | 3 |
| Nicotiana tabacum | Tobacco | 5 | 0 | 3 |
| Brassica oleracea | Broccoli | 4 | 0 | 0 |
| Pisum sativum | Pea | 4 | 1 | 0 |
| Glycine max | Soybean | 4 | 0 | 1 |
| Sorghum bicolor | Sorghum | 4 | 0 | 0 |
| Solanum tuberosum | Potato | 3 | 0 | 3 |
| Zea mays | Maize | 3 | 0 | 0 |
| Hordeum vulgare | Barley | 3 | 0 | 0 |
| Astragalus sinicus | Chinese Milk Vetch | 1 | 0 | 1 |
| Chenopodium rubrum | Red Goosefoot | 1 | 1 | 0 |
| Festuca pratensis Huds. | Fescue | 1 | 1 | 0 |
| Ipomoea nil | Japanese Morning Glory | 1 | 0 | 1 |
| Medicago sativa | Alfalfa | 1 | 0 | 1 |
| Rosa hybrid | Rose | 1 | 0 | 0 |
| Triticum turgidum | Wheat | 1 | 0 | 1 |
| Triticum aestivum | Wheat | 1 | 0 | 0 |
| Total | 21 | 1145 | 96 | 58 |
The information of 154 leaf senescence related mutants such as name, ecotype and mutagenesis method were also retrieved from literatures. Expression profiling data were acquired from a classical and systemic research paper (5).
In addition to manual curation, computational approaches were also employed to annotate these SAGs. We predicted the potential miRNA targets for the SAGs using the RNAhybrid method (7). The orthologs of each SAG in other plants were retrieved from the online database OrthoMCL-DB (8). Finally, putative function domains of SAGs-encoding proteins were identified with InterProScan (9).
IMPLEMENTATION OF ANALYSIS TOOLS
To meet the general requirement of data analysis, we integrated the sequence similarity search tool BLAST (10) and the sequence analysis platform WebLab (6) into our leaf senescence database. Users can either retrieve the sequence from LSD, or upload their own sequences to search homologs against different divisions (gene, mRNA, CDS and protein) in the LSD. WebLab is a web-based bioinformatics platform developed by our center and publicly available worldwide (http://www.weblab.org.cn/) (6). More than 200 programs covering all aspects of sequence analysis were integrated into WebLab. A user space is provided to save input data and analysis results for registered users. Users may retrieve sequence data and submit to WebLab directly to perform extensive analysis for DNA, mRNA and protein sequences of the SAGs they are interested in.
BROWSING AND SEARCHING THE DATABASE ENTRIES
The LSD database enables users to retrieve and analyze SAGs through the Browse or Search page. Users may browse the entries by clicking the buttons of Species, Mutants or Phenotypes at the main page. A tree-like structure was designed for both species and phenotypes, and a table was created for mutants. Currently, the major source of SAGs was from the two model organisms A. thaliana (949 entries) and Oryza sativa (104 entries). The phenotypes of all SAGs were divided into the following groups: (i) natural senescence, (ii) dark induced senescence, (iii) nutrition deficiency induced senescence, (iv) stress induced senescence and (v) others. The text search interface allows users to make queries with three types of data: (i) locus name, GenBank ID, alias, species and description of genes; (ii) name, type and ecotype of mutants; and (iii) title, author, journal and date of literature papers.
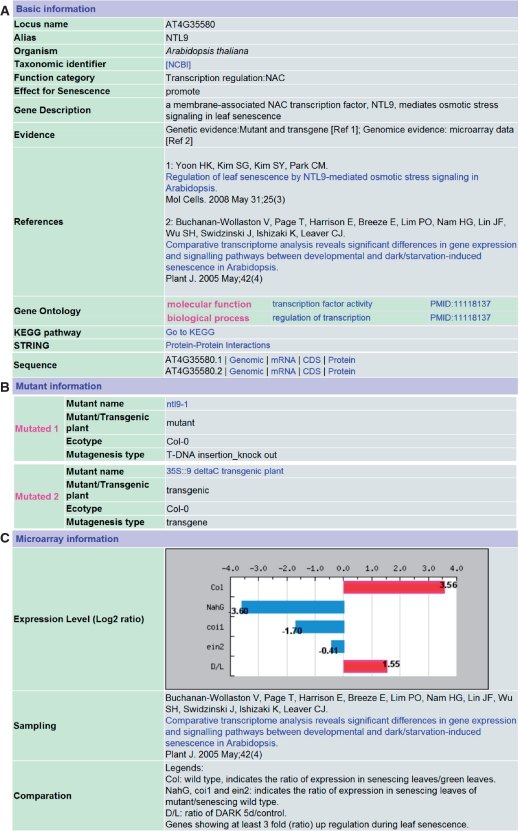
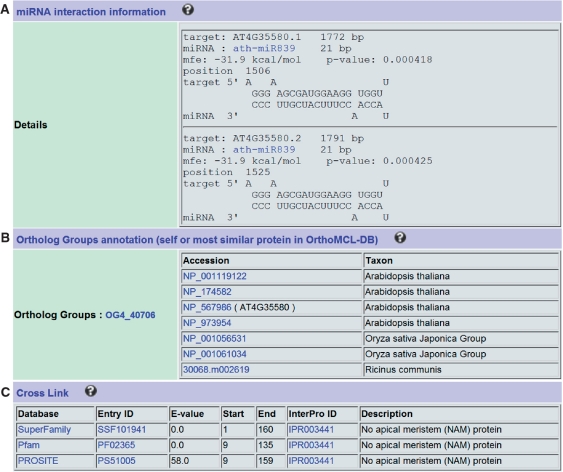
Figure 1 shows the annotation for a typical LSD entry NTL9, a member of the NAC transcription factor family and a membrane-associated gene that mediates osmotic stress signaling in leaf senescence (11). General information such as locus name, alias, organism, taxonomy was retrieved from the literature. Functional category, effect and evidence of senescence, as well as a brief description of this gene were manually annotated (Figure 1A). Mutant information is also provided (Figure 1B). Expression profiles generated from microarray data can be found for most Arabidopsis SAGs (Figure 1C). We predicted potential miRNA targets for some SAGs and added links to miRBase (12) for these miRNAs (Figure 2A). Orthologs from other plants are listed with links to the OrthoMCL database (Figure 2B). Putative functional domains of proteins encoded by SAGs were identified and annotated using the InterProScan program (9), and matches were displayed with cross links to several protein domain and family databases such as Prosite and Pfam (Figure 2C).
Figure 1.
A typical entry in LSD, the Arabidopsis NAC transcription factor. (A) Basic information, (B) Mutant information and (C) Expression Profile.
Figure 2.
Computational annotations for the Arabidopsis NAC transcription factor. (A) miRNA targets, (B) Ortholog Groups and (C) Cross Links to other databases.
ANALYZING THE SEQUENCE DATA
In addition to sequence similarity search with the BLAST tool kit implanted in the database, users may also perform extensive analysis for sequence data retrieved from the LSD. For each entry, links to different sequence types (Genomic, mRNA, CDS and Protein) are provided. Users can click these links to bring up the corresponding sequence and submit it to WebLab for further analysis, such as predicting gene structures, making pairwise or multiple sequence alignment, generating sequence logos, constructing phylogenetic trees, finding sequence motifs, etc.
FUTURE PLANS
The entries of LSD provided in this first release represent a preliminary data set. We will update the database regularly with more leaf senescence related data available, and predict putative SAGs from completely sequenced plant genomes in the near future. We will improve the user interface with comments and suggestions from the user community and add more documents including case studies to help user to make thorough analysis of SAGs. We hope that LSD can be a platform not only for the domestic and international collaborators we are working with, but also for the research community of leaf senescence worldwide.
FUNDING
National Natural Science Foundation of China (31071160 to J.L., 30625003 and 30730011 to H.G.); Ministry of Science and Technology of China (2009CB119101 to H.G.); Ministry of Education of China (NGI2008-108-3 to J.L., ED20060047 to H.G.). Funding for open access charge: National Laboratory of Protein Engineering and Plant Genetic Engineering.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank W.R. He and X. Tang for the design of the LSD web logo. We appreciate the anonymous reviewers for their valuable comments and suggestions.
REFERENCES
- 1.Lim PO, Kim HJ, Nam HG. Leaf senescence. Annu. Rev. Plant Biol. 2007;58:115–136. doi: 10.1146/annurev.arplant.57.032905.105316. [DOI] [PubMed] [Google Scholar]
- 2.Guo Y, Gan S. Leaf senescence: signals, execution, and regulation. Curr. Top. Dev. Biol. 2005;71:83–112. doi: 10.1016/S0070-2153(05)71003-6. [DOI] [PubMed] [Google Scholar]
- 3.Lim PO, Nam HG. The molecular and genetic control of leaf senescence and longevity in Arabidopsis. Curr. Top. Dev. Biol. 2005;67:49–83. doi: 10.1016/S0070-2153(05)67002-0. [DOI] [PubMed] [Google Scholar]
- 4.Rivero RM, Kojima M, Gepstein A, Sakakibara H, Mittler R, Gepstein S, Blumwald E. Delayed leaf senescence induces extreme drought tolerance in a flowering plant. Proc. Natl Acad. Sci. USA. 2007;104:19631–19636. doi: 10.1073/pnas.0709453104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Buchanan-Wollaston V, Page T, Harrison E, Breeze E, Lim PO, Nam HG, Lin JF, Wu SH, Swidzinski J, Ishizaki K, et al. Comparative transcriptome analysis reveals significant differences in gene expression and signalling pathways between developmental and dark/starvation-induced senescence in Arabidopsis. Plant J. 2005;42:567–585. doi: 10.1111/j.1365-313X.2005.02399.x. [DOI] [PubMed] [Google Scholar]
- 6.Liu X, Wu J, Wang J, Liu X, Zhao S, Li Z, Kong L, Gu X, Luo J, Gao G. WebLab: a data-centric, knowledge-sharing bioinformatic platform. Nucleic Acids Res. 2009;37:W33–W39. doi: 10.1093/nar/gkp428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Alves-Junior L, Niemeier S, Hauenschild A, Rehmsmeier M, Merkle T. Comprehensive prediction of novel microRNA targets in Arabidopsis thaliana. Nucleic Acids Res. 2009;37:4010–4021. doi: 10.1093/nar/gkp272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Chen F, Mackey AJ, Stoeckert CJ, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Zdobnov EM, Apweiler R. InterProScan - an integration platform for the signature-recognition methods in InterPro. Bioinformatics. 2001;17:847–848. doi: 10.1093/bioinformatics/17.9.847. [DOI] [PubMed] [Google Scholar]
- 10.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 11.Yoon HK, Kim SG, Kim SY, Park CM. Regulation of leaf senescence by NTL9-mediated osmotic stress signaling in Arabidopsis. Mol. Cells. 2008;25:438–445. [PubMed] [Google Scholar]
- 12.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]