Abstract
MPromDb (Mammalian Promoter Database) is a curated database that strives to annotate gene promoters identified from ChIP-seq results with the goal of providing an integrated resource for mammalian transcriptional regulation and epigenetics. We analyzed 507 million uniquely aligned RNAP-II ChIP-seq reads from 26 different data sets that include six human cell-types and 10 distinct mouse cell/tissues. The updated MPromDb version consists of computationally predicted (novel) and known active RNAP-II promoters (42 893 human and 48 366 mouse promoters) from various data sets freely available at NCBI GEO database. We found that 36% and 40% of protein-coding genes have alternative promoters in human and mouse genomes and ∼40% of promoters are tissue/cell specific. The identified RNAP-II promoters were annotated using various known and novel gene models. Additionally, for novel promoters we looked into other evidences—GenBank mRNAs, spliced ESTs, CAGE promoter tags and mRNA-seq reads. Users can search the database based on gene id/symbol, or by specific tissue/cell type and filter results based on any combination of tissue/cell specificity, Known/Novel, CpG/NonCpG, and protein-coding/non-coding gene promoters. We have also integrated GBrowse genome browser with MPromDb for visualization of ChIP-seq profiles and to display the annotations. The current release of MPromDb can be accessed at http://bioinformatics.wistar.upenn.edu/MPromDb/.
INTRODUCTION
The mammalian transcriptome and proteome is far more diverse than expected from one gene→one mRNA→one protein paradigm (1). This diversity arises due to the generation of multiple transcripts from a gene using alternative transcriptional and splicing events. Alternative transcriptional events that involve use of multiple promoters and/or transcriptional termination result in multiple pre-mRNAs from the same gene that can further undergo alternative splicing to generate a plethora of transcript variants corresponding to a single gene (2). Therefore, a gene can yield transcript variants that differ in either their regulatory UTRs or/and protein coding regions; thereby expanding the complexity of mammalian genomes (3–5). In particular, the role of alternative promoter activity is critical in transcriptional regulation, as their precise utilization allows the balanced expression of corresponding pre-mRNA variants in different cell and/or developmental contexts. In fact, recent evidence suggests that at least half of the mammalian genes use alternative promoters generating multiple transcript variants (3,5). Therefore, identifying all possible gene promoters, their usage and epigenetic modification states in specific cell populations, tissues and their developmental stages and disease conditions is critical to understanding a diversity of physiological processes associated with normal and diseased states.
Several high-throughput technologies, such as cap analysis gene expression (CAGE), chromatin immunoprecipitation (ChIP) followed by microarray analysis (ChIP–chip), (6,7), and more recently, ChIP coupled with sequencing (ChIP-seq) (8) and sequencing of cDNAs (RNA-seq) (5), are enabling the genome-wide identification of alternative promoters and their patterns of use. However, these high-throughput approaches need to be applied with caution because of the inherent problems with each method (9). In our recent study, we have shown that a combination of ChIP-seq and computational technique provides a better approach to annotate active promoters (9,10). Although EPD database (11) provides curated promoter sequences for eukaryotic organisms, it does not provide promoter activity information at tissue/cell centric level. In this update of MPromDb we have removed ChIP–chip results and added active RNAP-II promoters identified after analyzing six different cell types of human and 10 different cell/tissue types of mouse ChIP-seq experiments performed with RNAP-II antibody. In addition, we have added enrichment profile of various transcription factors obtained from ChIP-seq data sets. These promoters along with their annotations are provided as a user-friendly database, where each known and ChIP-seq promoter is linked to a new interface for visualization of enrichment profile. Here, we describe the updates of our MPromDb, which enables users to study promoter activity at tissue/cell centric level for human and mouse genome.
NEW FEATURES
Statistics of the promoters identified using ChIP-seq data sets
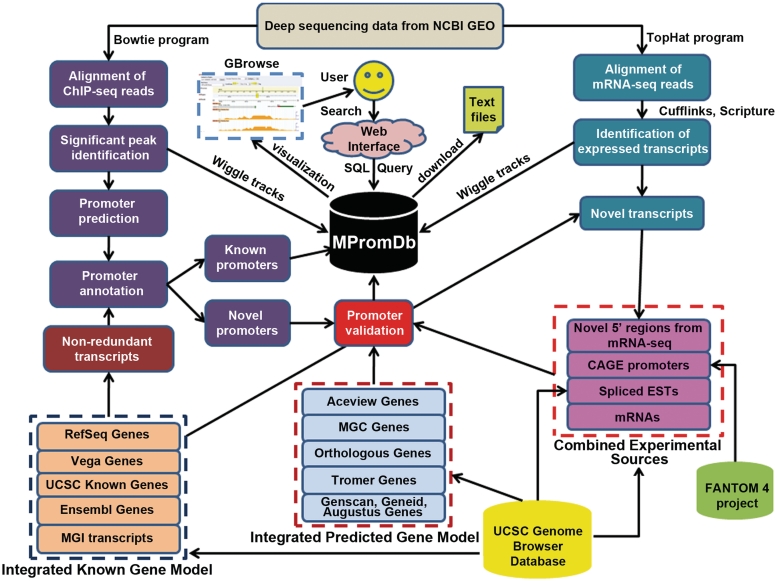
In this update, we have added (i) a comprehensive knowledgebase of known and novel promoters, (ii) promoters identified from RNAP-II ChIP-seq experiments, (iii) advance search and filter options and (iv) visualization of ChIP-seq profiles and promoters using GBrowse (12). The comprehensive promoter knowledgebase was generated from various known gene models (RefSeq, Vega, Ensembl, MGI and UCSC Known genes), predicted gene models (AceView, Tromer, MGC, SGP, SIB, Genscan, Geneid, N-SCAN and Augustus Abinitio), Orthologous gene model (XenoRef), GenBank mRNAs, spliced ESTs, CAGE promoters and mRNA-seq tags (Figure 1). The gene models, mRNAs and spliced ESTs were downloaded from UCSC Genome Browser database (13), CAGE promoters location were downloaded from FANTOM4 project (14) and mRNA-seq raw reads were downloaded from NCBI GEO database. We have also added promoter regions of recently discovered non-coding genes class (lincRNA) transcribed by RNAP-II (15,16). The total number of records in the knowledgebase can be found in Table S1.
Figure 1.
The block diagram and workflow of updated MPromDb database. Deep sequencing datasets were downloaded from NCBI GEO server and processed by our analysis and annotation pipeline. The identified promoters are deposited in MPromDb tables. Novel promoters are compared to various existing experimental and predicted gene promoter regions and status of novel promoters is deposited in the relational tables. The database is accessed through a user-friendly webpage. The database is integrated with open source genome browser (GBrowse) to visualize the promoter and various ChIP-seq enrichment profiles.
The RNAP-II ChIP-seq data sets includes the data generated at our lab (9) and data sets from various published and unpublished studies available freely at NCBI GEO database. The human RNAP-II ChIP-seq data sets include six different cell lines: CD4 + T, HeLa S3, K562, NB4, Lymphoblastoid and Jurkat, whereas mouse samples include five different tissues and five different cell types: brain, liver, lung, spleen, kidney, Embryonic Stem Cell (V6.5), Mouse Embryonic Fibroblasts B4, Mouse Embryonic Fibroblasts B6, Bone Marrow-derived macrophages and 3T3-L1 (9,17–23). The NCBI GEO accession numbers of the data sets are provided in Table S2. On the downloaded ChIP-seq data sets, we apply our pipeline (Figure 1) that includes alignment, identification of significant enriched regions, promoter prediction and annotation. Bowtie program (24) was applied to map reads to the reference genome (mm9 version for mouse and hg18 version for human), allowing up to two mismatches. Only uniquely mapped reads were considered for further analysis. We obtained 174 777 943 and 333 192 049 uniquely mapped reads for mouse and human genome respectively (Table S3). Significant peaks were identified using our three steps procedure as described in (9) at P-value = 0.01. After identification of significant RNAP-II bound peaks we apply our recently published program for prediction of RNAP-II bound promoters (10). The peak identification and promoter prediction of each sample is summarized in Table S3. Following promoter prediction, we performed promoter annotation using our reference promoter knowledgebase as summarized in Figures S1 and S2. Finally, we identified 48 366 mouse and 42 893 human promoters bound by RNAP-II where 39% and 42% of the promoters in mouse and human respectively were annotated as ‘Novel promoters’ (Table 1). In case the predicted ChIP-seq promoters lie within −1 to 0.5 kb of known TSS or within the first exons of known transcripts, they are defined as ‘Known promoters’ otherwise they are considered as ‘Novel promoters’. It is worth noting that 65% and 90% of novel promoters in mouse and human, respectively, are supported by additional sources (novel gene models, mRNAs, spliced ESTs, CAGE tags and Orthologous gene model) (Table S4).
Table 1.
Summary of RNAP-II bound promoters identified in various tissues/cell types for human and mouse using ChIP-seq data sets
| Species | Tissue/cell type | No. of known promoters | No. of novel promoters | No. of tissue/ cell-specific promoters | No. of CpG promoters | No. of bidirectional promoters | No. of total promoters |
|---|---|---|---|---|---|---|---|
| Mouse | Brain | 15 948 | 5270 | 3978 | 13 864 | 1373 | 21 218 |
| Liver | 12 319 | 3189 | 1642 | 10 421 | 1250 | 15 508 | |
| Kidney | 15 059 | 4632 | 1995 | 12 879 | 1348 | 19 691 | |
| Spleen | 9089 | 2121 | 806 | 8273 | 1067 | 11 210 | |
| Lung | 15 373 | 5142 | 1935 | 13 986 | 1374 | 20 515 | |
| Embryonic stem cell(V6.5) | 11 895 | 2880 | 2745 | 12 063 | 1314 | 14 775 | |
| Mouse embryonic fibroblasts B4 | 10 558 | 2261 | 273 | 10 898 | 1241 | 12 819 | |
| Mouse embryonic fibroblasts B6 | 11 887 | 2761 | 706 | 10 886 | 1237 | 14 648 | |
| Bone marrow-derived macrophages (untreated) | 13 320 | 3977 | 870 | 12 038 | 1298 | 17 297 | |
| Bone marrow-derived macrophages (2 h) | 12 647 | 3713 | 566 | 11 846 | 1294 | 16 260 | |
| Bone marrow-derived macrophages (4 h) | 13 119 | 4041 | 688 | 11 926 | 1292 | 17 160 | |
| 3T3-L1 cells (untreated) | 8489 | 1373 | 113 | 8597 | 1038 | 9862 | |
| 3T3-L1 cells (Day 1) | 8684 | 1626 | 154 | 8803 | 1072 | 9310 | |
| 3T3-L1 cells (Day 2) | 8508 | 1593 | 174 | 8415 | 1042 | 10 101 | |
| 3T3-L1 cells (Day 3) | 8374 | 1540 | 136 | 8371 | 1035 | 9914 | |
| 3T3-L1 cells (Day 4) | 6976 | 1422 | 194 | 6793 | 848 | 8398 | |
| 3T3-L1 cells (Day 6) | 4039 | 1443 | 927 | 4030 | 511 | 5482 | |
| Total | 29 517 | 18 849 | 17 902 | 24 587 | 1801 | 48 366 | |
| Human | Jurkat cells | 7417 | 1403 | 541 | 7653 | 792 | 8820 |
| K562 cells | 16 410 | 8012 | 6422 | 16 918 | 1320 | 24 422 | |
| Lymphoblastoid cells | 19 617 | 8998 | 6629 | 20 682 | 1311 | 28 615 | |
| NB4 cells | 12 925 | 2944 | 916 | 13 650 | 1156 | 15 869 | |
| HeLa_S3 cells | 13 982 | 3502 | 2101 | 14 812 | 1212 | 17 484 | |
| CD4 + T cells | 14 329 | 4137 | 1336 | 15 740 | 1220 | 18 466 | |
| CD4 + T cells (2 h) | 7354 | 1267 | 174 | 7955 | 882 | 8621 | |
| CD4 + T cells (12 h) | 11 470 | 2389 | 377 | 12 740 | 1172 | 13 859 | |
| Total | 24 967 | 17 926 | 18 496 | 27 488 | 1501 | 42 893 |
Furthermore, our analysis has identified promoters for 15 493 and 14 266 protein-coding genes in mouse and human respectively. A gene is defined as protein coding if it has at least one protein-coding transcript in RefSeq/Vega gene models, or else it is a non-coding gene. Please note that a protein coding gene can generate transcript variants that are non-coding RNAs. We also observed that 40% and 36% of protein coding genes in mouse and human are expressed from alternative promoters (Table 2). Surprisingly, 37% of promoters in mouse and 43% of human promoters were identified in a single cell/tissue suggesting that they are cell/tissue-specific promoters. Additionally, we analyzed the CpG-richness and bidirectionality of the promoters and found that 51% and 64% of promoters are CpG-rich and there are 1801 and 1501 bidirectional promoters in mouse and human respectively. Additionally, we also provide significant enrichment profiles of various factors (Mouse – OCT4, CEBPa, CHD7, c-Myc, CTCF, ESRRB, FOXA1, FOXA2, GFP, KLF4, n-Myc, NR5A2, P300, Rbbp5, SETDB1, SIRT1, SOX2, STAT3, STAT4, STAT6, SUZ12, TBP, TBX3, TCFCP2I1, WDR5, ZFX; Human – OCT4, CBP, CTCF, ETS1, KLF4, NANOG, P300, PCAF, PHF8, PPARG, RUNX, SOX2, STAT1, TFII, Tip60, ZNF263, SUZ12, MOF, IGF1R, NFkB) calculated from different published and unpublished ChIP-seq datas ets (Table S5A and B).
Table 2.
Alternative promoter usage for active protein-coding genes in mouse and human
| Protein-coding genes | Mouse (%) | Human (%) |
|---|---|---|
| 1-promoter genes | 9290 (60) | 9051 (63.44) |
| 2-promoter genes | 3490 (22.5) | 3192 (22.37) |
| ≥3-promoter genes | 2707 (17.5) | 2023 (14.18) |
| Total | 15 493 | 14 266 |
Database search and visualization
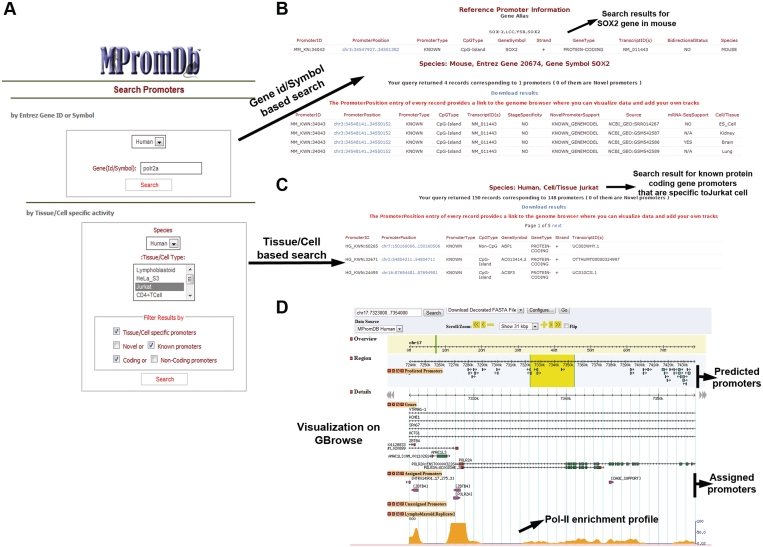
MPromDb as a web-based application has many layers: the core application (designed in Django), a backend database (MySQL), a visualization component (GBrowse) and a web server (Apache) (see Supplementary File 1). The promoter information corresponding to a particular gene can be retrieved from the database using Entrez geneid or gene symbol. We also provide additional search and filter options such as selection of tissue/cell type, tissue/cell specific promoters, known/novel promoters and coding/non-coding gene promoters. The gene search query returns result at two different levels (see Figure 2, Supplementary File 2, Supplementary Tables S6 and S7). The first level provides information (promoter position, CpG type and bidirectional type) regarding all promoters of the queried gene that are present in the promoter knowledgebase. The second level of search result lists all promoters identified from ChIP-seq data sets for the queried gene. The result of the search can be downloaded into an excel file. Each promoter of the search result is linked to the visualization module. Further, complete list of annotated promoters can be downloaded from the download link. Visualization of the promoter position and ChIP-seq data enrichment profile is implemented using GBrowse (12), an open source genome browser platform. GBrowse is simple but highly configurable web-based genome browser, which provides a fast and customizable interface for visualizing data that is stored in a backend database, as well as the data that is uploaded by the user. GBrowse is lighter than UCSC genome browser and offers many advantages especially in displaying the results and tracks. Some of the features unique to GBrowse are: glyphs and balloons to represent different features, organizing features sub categories to more depth, multi-language support, view GenBank, chado and biosql feature databases, third party loading. On GBrowse the identified promoter location and enrichment profile of the analyzed ChIP-seq data sets are shown (Figure 2D). Further, users can directly type the genome coordinates or gene symbol on GBrowse for searching. Users have an option to turn on/off the tracks that are displayed on the genome browser.
Figure 2.
Screenshots of MPromDb and search results. (A) MPromDb main search page where a user can perform search based on either Entrez gene id/symbol or specific tissue/cell type and the resulting page is shown in (B) and (C), respectively. (D) User can visualize the ChIP-seq profile for any promoter displayed on (B) or (C) by clicking on the promoter position link.
FUTURE PLANS
In future, we plan to include epigenetic histone modifications profile identified from ChIP-seq data sets that are currently available at NCBI GEO and integrate it to our promoter knowledgebase. We will also continue to collect RNAP-II and transcription factors ChIP-seq data sets from a wider variety of tissues and cell types to routinely update MPromDb. We also plan to include other mammalian data sets, and add additional features and search options to the frontend of the database. In conclusion, MPromDb will provide integrated transcriptional regulatory information for mammalian genomes in an easily accessible way. We believe that the updates will facilitate large-scale ChIP-seq data analysis and contribute toward the elucidation of mammalian transcriptional regulatory networks.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
NHGRI/NIH grant (# R01HG003362); American Cancer Society Research Scholar Grant (# RSG-07-097-01 to R.D.); and Philadelphia Healthcare Trust. R.D. holds a Philadelphia Healthcare Trust Endowed Chair Position. Funding for open access charge: National Institutes of Health grant (#R01HG003362 to R.D.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Sharmistha Pal for reading the manuscript and providing valuable inputs for developing MPromDb. The use of computational resources in the Centre for Systems and Computational Biology and Bioinformatics Facility of Wistar Cancer Centre (grant # P30CA010815) are gratefully acknowledged.
REFERENCES
- 1.Moore MJ, Proudfoot NJ. Pre-mRNA processing reaches back to transcription and ahead to translation. Cell. 2009;136:688–700. doi: 10.1016/j.cell.2009.02.001. [DOI] [PubMed] [Google Scholar]
- 2.Hallegger M, Llorian M, Smith CW. Alternative splicing: global insights. FEBS J. 2010;277:856–866. doi: 10.1111/j.1742-4658.2009.07521.x. [DOI] [PubMed] [Google Scholar]
- 3.Pan Q, Shai O, Lee LJ, Frey BJ, Blencowe BJ. Deep surveying of alternative splicing complexity in the human transcriptome by high-throughput sequencing. Nat. Genet. 2008;40:1413–1415. doi: 10.1038/ng.259. [DOI] [PubMed] [Google Scholar]
- 4.Kimura K, Wakamatsu A, Suzuki Y, Ota T, Nishikawa T, Yamashita R, Yamamoto J, Sekine M, Tsuritani K, Wakaguri H, et al. Diversification of transcriptional modulation: large-scale identification and characterization of putative alternative promoters of human genes. Genome Res. 2006;16:55–65. doi: 10.1101/gr.4039406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Wang ET, Sandberg R, Luo S, Khrebtukova I, Zhang L, Mayr C, Kingsmore SF, Schroth GP, Burge CB. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456:470–476. doi: 10.1038/nature07509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sandelin A, Carninci P, Lenhard B, Ponjavic J, Hayashizaki Y, Hume DA. Mammalian RNA polymerase II core promoters: insights from genome-wide studies. Nat. Rev. Genet. 2007;8:424–436. doi: 10.1038/nrg2026. [DOI] [PubMed] [Google Scholar]
- 7.Kapranov P, Willingham AT, Gingeras TR. Genome-wide transcription and the implications for genomic organization. Nat. Rev. Genet. 2007;8:413–423. doi: 10.1038/nrg2083. [DOI] [PubMed] [Google Scholar]
- 8.Robertson G, Hirst M, Bainbridge M, Bilenky M, Zhao Y, Zeng T, Euskirchen G, Bernier B, Varhol R, Delaney A, et al. Genome-wide profiles of STAT1 DNA association using chromatin immunoprecipitation and massively parallel sequencing. Nat. Methods. 2007;4:651–657. doi: 10.1038/nmeth1068. [DOI] [PubMed] [Google Scholar]
- 9.Sun H, Wu J, Wickramasinghe P, Pal S, Gupta R, Bhattacharya A, Agosto-Pérez FJ, Showe LC, Huang TH, Davuluri RV. Genome-wide mapping of RNA Pol-II promoter usage in mouse tissues by ChIP-seq. Nucleic Acids Res. 2010 doi: 10.1093/nar/gkq775. doi:10.1093/nar/gkq775 [Epub ahead of print 14 September 2010]. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Gupta R, Wikramasinghe P, Bhattacharyya A, Perez FA, Pal S, Davuluri RV. Annotation of gene promoters by integrative data-mining of ChIP-seq Pol-II enrichment data. BMC Bioinformatics. 2010;11(Suppl. 1):S65. doi: 10.1186/1471-2105-11-S1-S65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schmid CD, Perier R, Praz V, Bucher P. EPD in its twentieth year: towards complete promoter coverage of selected model organisms. Nucleic Acids Res. 2006;34:D82–D85. doi: 10.1093/nar/gkj146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Severin J, Waterhouse AM, Kawaji H, Lassmann T, van Nimwegen E, Balwierz PJ, de Hoon MJ, Hume DA, Carninci P, Hayashizaki Y, et al. FANTOM4 EdgeExpressDB: an integrated database of promoters, genes, microRNAs, expression dynamics and regulatory interactions. Genome Biol. 2009;10:R39. doi: 10.1186/gb-2009-10-4-r39. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Guttman M, Amit I, Garber M, French C, Lin MF, Feldser D, Huarte M, Zuk O, Carey BW, Cassady JP, et al. Chromatin signature reveals over a thousand highly conserved large non-coding RNAs in mammals. Nature. 2009;458:223–227. doi: 10.1038/nature07672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Guttman M, Garber M, Levin JZ, Donaghey J, Robinson J, Adiconis X, Fan L, Koziol MJ, Gnirke A, Nusbaum C, et al. Ab initio reconstruction of cell type-specific transcriptomes in mouse reveals the conserved multi-exonic structure of lincRNAs. Nat. Biotechnol. 2010;28:503–510. doi: 10.1038/nbt.1633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wang Z, Zang C, Cui K, Schones DE, Barski A, Peng W, Zhao K. Genome-wide mapping of HATs and HDACs reveals distinct functions in active and inactive genes. Cell. 2009;138:1019–1031. doi: 10.1016/j.cell.2009.06.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barski A, Jothi R, Cuddapah S, Cui K, Roh TY, Schones DE, Zhao K. Chromatin poises miRNA- and protein-coding genes for expression. Genome Res. 2009;19:1742–1751. doi: 10.1101/gr.090951.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kasowski M, Grubert F, Heffelfinger C, Hariharan M, Asabere A, Waszak SM, Habegger L, Rozowsky J, Shi M, Urban AE, et al. Variation in transcription factor binding among humans. Science. 328:232–235. doi: 10.1126/science.1183621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Mikkelsen TS, Ku M, Jaffe DB, Issac B, Lieberman E, Giannoukos G, Alvarez P, Brockman W, Kim TK, Koche RP, et al. Genome-wide maps of chromatin state in pluripotent and lineage-committed cells. Nature. 2007;448:553–560. doi: 10.1038/nature06008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Seila AC, Calabrese JM, Levine SS, Yeo GW, Rahl PB, Flynn RA, Young RA, Sharp PA. Divergent transcription from active promoters. Science. 2008;322:1849–1851. doi: 10.1126/science.1162253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.De Santa F, Narang V, Yap ZH, Tusi BK, Burgold T, Austenaa L, Bucci G, Caganova M, Notarbartolo S, Casola S, et al. Jmjd3 contributes to the control of gene expression in LPS-activated macrophages. EMBO J. 2009;28:3341–3352. doi: 10.1038/emboj.2009.271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Nielsen R, Pedersen TA, Hagenbeek D, Moulos P, Siersbaek R, Megens E, Denissov S, Borgesen M, Francoijs KJ, Mandrup S, et al. Genome-wide profiling of PPARgamma:RXR and RNA polymerase II occupancy reveals temporal activation of distinct metabolic pathways and changes in RXR dimer composition during adipogenesis. Genes Dev. 2008;22:2953–2967. doi: 10.1101/gad.501108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Langmead B, Trapnell C, Pop M, Salzberg SL. Ultrafast and memory-efficient alignment of short DNA sequences to the human genome. Genome Biol. 2009;10:R25. doi: 10.1186/gb-2009-10-3-r25. [DOI] [PMC free article] [PubMed] [Google Scholar]