Abstract
PCDB (http://www.pcdb.unq.edu.ar) is a database of protein conformational diversity. For each protein, the database contains the redundant compilation of all the corresponding crystallographic structures obtained under different conditions. These structures could be considered as different instances of protein dynamism. As a measure of the conformational diversity we use the maximum RMSD obtained comparing the structures deposited for each domain. The redundant structures were extracted following CATH structural classification and cross linked with additional information. In this way it is possible to relate a given amount of conformational diversity with different levels of information, such as protein function, presence of ligands and mutations, structural classification, active site information and organism taxonomy among others. Currently the database contains 7989 domains with a total of 36581 structures from 4171 different proteins. The maximum RMSD registered is 26.7 Å and the average of different structures per domain is 4.5.
INTRODUCTION
Protein conformational diversity is a key feature to understand protein function. Since the early studies of Max Perutz on the T and R forms of hemoglobin, increasing experimental evidence supports the notion that native state of proteins is not unique. In fact, the native state is better represented by an ensemble of conformers in equilibrium describing the conformational diversity or dynamism of a protein (1). It has been showed that the ensemble description is essential to understand central biological aspects of protein function such as the catalytic process of enzymes (2–4), protein–protein recognition (5–7), macromolecular process such as DNA replication and protein folding by chaperonins (8), enzyme promiscuity (9), signal transduction (10,11) and the proteins ability to develop new functions (property known as ‘evolvability’) (12,13). Despite that, the characterization of the equilibrium ensemble of conformers, involving the study of the structural and thermodynamic features of each individual conformer, represents a major challenge to overcome. In this way, different procedures have been applied to the study of protein dynamisms. Experimentally, the nuclear magnetic resonance (NMR) spectroscopy is among the most widely used approaches representing a promising and active area of research (14). On the other hand, computational methods like Coarse-Grained Molecular Dynamics and Monte Carlo methods techniques, used in combination with Normal mode analysis, have been revealed that they are useful tools to explore the conformational landscape of proteins (15–19). Finally, a completely different approach to study conformational diversity considers that crystallographic structures of the same protein obtained under different conditions are snapshots or instances of protein dynamism. This view is supported by the correlation found between the observed structural diversity determined by solution experiments such as NMR measurements and those coming from crystallographic structures of proteins obtained in different conditions (6,20–25). Also a good correlation was found when computational methods, such as molecular dynamics, were used to simulate protein dynamism and then compared with solution structures from NMR (26,27).
With thousands of structures redundantly deposited in structural databases (28) the extension and distribution of the conformational diversity can be explored for a large number of proteins not accessible with the methodologies mentioned above. In this paper we have used this approach to develop a database of proteins with conformational diversity. Here, we describe PCDB (from protein Conformational diversity database), its web functionality and possible applications.
OVERVIEW OF PCDB
PCDB is a database of proteins showing conformational diversity. As was mentioned above, conformational diversity is estimated from a redundant collection of structures for each protein domain deposited in the database. PCDB was developed from CATH database v3.3 following its protein domain structural hierarchy and definitions (29). Briefly, CATH clusters proteins domains using structural and sequence similarities in a hierarchy defined by 9 levels called CATHSOLID where the ‘D’ level assigns a number for each individual domain in the database and corresponds with the collection of different crystallographic structures for an individual protein. This level was used to build PCDB collecting all the proteins domains with at least two different crystallographic structures classified in CATH. The current version of the PCDB contains 7989 protein domains from 4171 proteins and 34775 crystallographic structures and 1806 corresponding to NMR (Table 1).
Table 1.
Summary of the data available in PCDB v1.0
| Domains | 7989 |
| Proteins | 4171 |
| Structures | 36581 |
| Maximum RMSDmax | 26.7 |
| Average structures per domain | 4.7 |
| Structures in class mainly alphaa | 1728 |
| Structures in class mainly betaa | 2326 |
| Structures in class mixed alpha-betaa | 3790 |
| Structures in class few secondary structurea | 145 |
aAccordingly to CATH v3.3 (http://www.cathinfo.db) hierarchy (29).
The structures collected for each protein domain could have been crystallized under the same or different conditions. If the conditions were the same, it is known that RMSD between different structures is as much as 0.1 to 0.4 A (30). Larger RMSD are expected when conformational diversity appears and this could happen when crystallization conditions varies among the structures considered. In fact RMSD as high as 23.4 have been reported in redundant studies of protein structures (28). Following the addition of ligands, for example, it is well established that a conformational equilibrium shift towards a high affinity conformer could occurred originating changes in tertiary structure (12,31,32). Besides, other changes in crystallization conditions like modifications in the oligomerization state (33), pH and temperature, as well as the presence of mutations (34) can also modify the relative stability of conformers and then originate differences between crystallographic structures for the same protein. In addition, different sequence modifications or crystallographic errors could introduce conformational diversity unrelated to biological reasons. Considering that our method to measure conformational diversity relies in the quality of the crystallographic structure, different filters were used in order to build the database. The different criteria used to select the structures are explained below and a general PCDB building scheme can be found in Supplementary Figure S1.
In PCDB, the structures are linked with information contained in PDB concerning the crystallization procedure and supplementary data that could help to understand the occurrence of conformational diversity. The factors considered are: the presence of ligands, mutations, changes in the oligomeric state and pH. The maximum RMSD (RMSDmax) among the redundant structures of each protein domain is used to evaluate the extension of the structural change. Using the data in PCDB, we have found that at least one of these set of selected experimental features is involved in the 74% of all the domains (Table 2), and in the 60% of the domains with more than 0.4 RMSDmax. Besides the information provided for the crystallization procedure, each of the proteins deposited in PCDB was cross linked with different databases. In this way, a given extension of conformational diversity measured by RMSDmax can be related with diverse biological and structural information such as biological function [GO terms (35) and Enzyme Commission numbers(EC) (36)], structural classification [CATH (29)], taxonomy (NCBI taxonomic ID and genus and species names), metabolic pathways location, subcellular location, protein interactions, protein family, presence of characterized catalytic site [Catalytic Site Atlas (37)] and derived InterPro links (38).
Table 2.
Number of proteins in PCDB with different factors possibly promoting the observed conformational diversity
| Possible condition promoting conformational diversity | Number of proteins |
|---|---|
| Mutations | 268 |
| Ligands | 568 |
| Changes in oligomeric state | 536 |
| Changes in pH | 1029 |
| Mutations and Ligands | 77 |
| Mutations and changes in oligomeric state | 108 |
| Mutations and changes in pH | 213 |
| Ligands and changes in oligomeric state | 231 |
| Ligands and changes in pH | 387 |
| Changes in oligomeric state and pH | 613 |
| Four conditions | 269 |
PCDB is composed of a web application based on PHP language, connected with a MySQL database. The database includes information derived from numerous biological databases and online servers and data acquired from personal scripting and programs. PCDB search tool is based on dynamics SQL queries generated in PHP. PCDB browsing capability is based on SQL stored procedures that are executed dynamically, using PHP language. PCDB was built using the redundant structures from each protein domain collected from CATH v3.3 (39) (see Supplementary Figure S1). The structures belonging to each protein domain were structurally aligned using MAMMOTH (40) and the RMSDmax between conformers were registered. Information about crystallization conditions was extracted from PDBML/XML files, as well as the oligomeric state, presence of sequence modifications, mutations, deletion and missing residues. Post-translational modifications were extracted from the ‘Controlled vocabulary of post-translational modifications’ provided by Uniprot. Information about catalytic residues was extracted from Catalytic Site Atlas (37). Further biological information for each structure were extracted from different databases: PDB (30), SIFTS (http://www.ebi.ac.uk/msd/sifts/) and UniProt (41).
APPLICATIONS
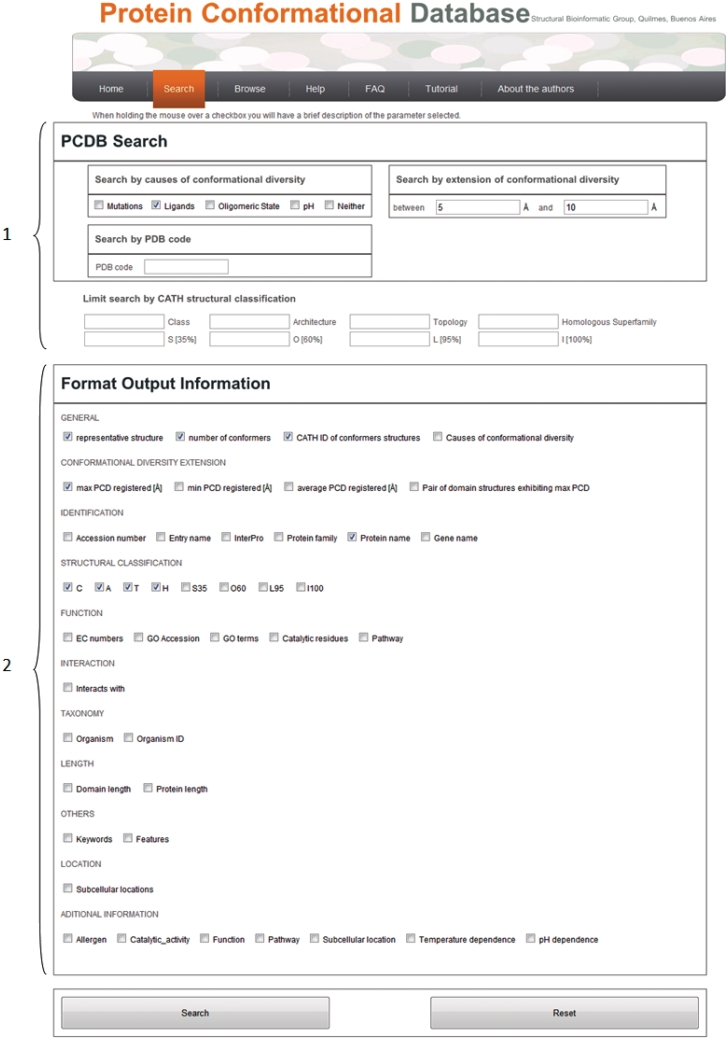
Conformational diversity is a central issue to understand protein function so its characterization could span multiple applications. PCDB database is designed to retrieve proteins with a given amount of conformational diversity measured by RMSDmax and allows relating this value with different levels of information. There are two main ways to explore PCDB (Figure 1). The main attribute to search PCDB concerns the extension of conformational diversity measured by RMSDmax. This type of search could be limited using a set of four attributes (presence of ligands, presence of mutations, changes in oligomerization state and changes in pH) considering the properties characterizing the experimental conditions of crystallization of each structure. These attributes can be selected separately or in different combinations (Table 2) and can be used to explain the RMSDmax obtained for a given protein. In the example showed in Figure 1, we were interested in searching PCDB for proteins with 5–10 RMSDmax between their respective structures due to the presence of ligands. Therefore, the resulted extension of conformational diversity can be univocally associated to conformational changes upon ligand binding. Also in Figure 1, and below the search field, the field to customize the output information is displayed. In this section it is possible to select different levels of information from structural classification, protein function or subcellular location among others. It is also possible to retrieve the structural superposition of the conformers with the maximum RMSD. Similar searches could explore PCBD using a single or a combination of the attributes producing conformational changes. Furthermore, the biological and structural data contained in the customizable output field, could be used to explore different trends related with conformational diversity.
Figure 1.
Searching PCDB using the presence of ligands as possible origin of conformational diversity between 5 and 10 units of RMSDmax (1). In the Format output section (2) it is possible to customize the biological and physicochemical information retrieved with the results.
FUTURE CONSIDERATIONS
We are interested in increasing the amount and diversity of available biological and structural data for each domain represented in the database, to enhance possible correlations studies between conformational diversity and a broad spectrum of physiochemical parameters. One of our near future goals is to introduce sequence alignments for each deposited protein to derive evolutionary information such as the relative conservation of different positions and evolutionary rates. The link between the pattern of residue substitution and the extension of conformational diversity is a promising field to increase our understanding about protein evolution; however it is almost an unexplored field yet. Beside this, and following previous works, we would like to enrich PCDB introducing structures from close homologous proteins (21) in order to increase the conformational representation of the deposited domains.
CONCLUSIONS
Two main features differentiate PCDB from other databases containing information about conformational diversity in proteins (42,43). Firstly, PCDB uses experimentally determined structures and secondly this data are related with biological and structural information to possible explains the observed conformational diversity extension. In the present version, PCDB contains 7989 protein domains with a broad range of conformational diversity from the trivial zero to 26.7 RMSDmax. In this way PCDB could be an essential tool to understand conformational diversity and by this means obtain a better understanding of protein function.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Proyectos de Investigacion Plurianuales (PIP) CONICET grant (112-200801-02849) and Universidad Nacional de Quilmes grant (53/B056); Ezequiel Juritz has a type II fellowship from CONICET. Funding for open access charge: CONICET and UNQ grants.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank the referees for all the helpful comments. Sebastian Fernandez Alberti and Gustavo Parisi are members of the Research Career of the National Research Council (CONICET, Argentina).
REFERENCES
- 1.Ma B, Kumar S, Tsai CJ, Nussinov R. Folding funnels and binding mechanisms. Protein Eng. 1999;12:713–720. doi: 10.1093/protein/12.9.713. [DOI] [PubMed] [Google Scholar]
- 2.Henzler-Wildman KA, Lei M, Thai V, Kerns SJ, Karplus M, Kern D. A hierarchy of timescales in protein dynamics is linked to enzyme catalysis. Nature. 2007;450:913–916. doi: 10.1038/nature06407. [DOI] [PubMed] [Google Scholar]
- 3.Henzler-Wildman KA, Thai V, Lei M, Ott M, Wolf-Watz M, Fenn T, Pozharski E, Wilson MA, Petsko GA, Karplus M, et al. Intrinsic motions along an enzymatic reaction trajectory. Nature. 2007;450:838–844. doi: 10.1038/nature06410. [DOI] [PubMed] [Google Scholar]
- 4.Wolf-Watz M, Thai V, Henzler-Wildman K, Hadjipavlou G, Eisenmesser EZ, Kern D. Linkage between dynamics and catalysis in a thermophilic-mesophilic enzyme pair. Nat. Struct. Mol. Biol. 2004;11:945–949. doi: 10.1038/nsmb821. [DOI] [PubMed] [Google Scholar]
- 5.Yogurtcu ON, Erdemli SB, Nussinov R, Turkay M, Keskin O. Restricted mobility of conserved residues in protein-protein interfaces in molecular simulations. Biophys. J. 2008;94:3475–3485. doi: 10.1529/biophysj.107.114835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Lange OF, Lakomek NA, Fares C, Schroder GF, Walter KF, Becker S, Meiler J, Grubmuller H, Griesinger C, de Groot BL. Recognition dynamics up to microseconds revealed from an RDC-derived ubiquitin ensemble in solution. Science. 2008;320:1471–1475. doi: 10.1126/science.1157092. [DOI] [PubMed] [Google Scholar]
- 7.Fuentes EJ, Der CJ, Lee AL. Ligand-dependent dynamics and intramolecular signaling in a PDZ domain. J. Mol. Biol. 2004;335:1105–1115. doi: 10.1016/j.jmb.2003.11.010. [DOI] [PubMed] [Google Scholar]
- 8.Russel D, Lasker K, Phillips J, Schneidman-Duhovny D, Velazquez-Muriel JA, Sali A. The structural dynamics of macromolecular processes. Curr. Opin. Cell Biol. 2009;21:97–108. doi: 10.1016/j.ceb.2009.01.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Khersonsky O, Roodveldt C, Tawfik DS. Enzyme promiscuity: evolutionary and mechanistic aspects. Curr. Opin. Chem. Biol. 2006;10:498–508. doi: 10.1016/j.cbpa.2006.08.011. [DOI] [PubMed] [Google Scholar]
- 10.Bai F, Branch RW, Nicolau DV, Jr, Pilizota T, Steel BC, Maini PK, Berry RM. Conformational spread as a mechanism for cooperativity in the bacterial flagellar switch. Science. 2010;327:685–689. doi: 10.1126/science.1182105. [DOI] [PubMed] [Google Scholar]
- 11.Smock RG, Gierasch LM. Sending signals dynamically. Science. 2009;324:198–203. doi: 10.1126/science.1169377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Tokuriki N, Tawfik DS. Protein dynamism and evolvability. Science. 2009;324:203–207. doi: 10.1126/science.1169375. [DOI] [PubMed] [Google Scholar]
- 13.Aharoni A, Gaidukov L, Khersonsky O, Mc QGS, Roodveldt C, Tawfik DS. The ‘evolvability’ of promiscuous protein functions. Nat. Genet. 2005;37:73–76. doi: 10.1038/ng1482. [DOI] [PubMed] [Google Scholar]
- 14.Lindorff-Larsen K, Best RB, Depristo MA, Dobson CM, Vendruscolo M. Simultaneous determination of protein structure and dynamics. Nature. 2005;433:128–132. doi: 10.1038/nature03199. [DOI] [PubMed] [Google Scholar]
- 15.Chng CP, Yang LW. Coarse-grained models reveal functional dynamics–II. Molecular dynamics simulation at the coarse-grained level: theories and biological applications. Bioinform. Biol. Insights. 2008;2:171–185. doi: 10.4137/bbi.s459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Karplus M, Kuriyan J. Molecular dynamics and protein function. Proc. Natl Acad. Sci. USA. 2005;102:6679–6685. doi: 10.1073/pnas.0408930102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tozzini V. Coarse-grained models for proteins. Curr. Opin. Struct. Biol. 2005;15:144–150. doi: 10.1016/j.sbi.2005.02.005. [DOI] [PubMed] [Google Scholar]
- 18.Bahar I, Rader AJ. Coarse-grained normal mode analysis in structural biology. Curr. Opin. Struct. Biol. 2005;15:586–592. doi: 10.1016/j.sbi.2005.08.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ma J. Usefulness and limitations of normal mode analysis in modeling dynamics of biomolecular complexes. Structure. 2005;13:373–380. doi: 10.1016/j.str.2005.02.002. [DOI] [PubMed] [Google Scholar]
- 20.Zoete V, Michielin O, Karplus M. Relation between sequence and structure of HIV-1 protease inhibitor complexes: a model system for the analysis of protein flexibility. J. Mol. Biol. 2002;315:21–52. doi: 10.1006/jmbi.2001.5173. [DOI] [PubMed] [Google Scholar]
- 21.Best RB, Lindorff-Larsen K, DePristo MA, Vendruscolo M. Relation between native ensembles and experimental structures of proteins. Proc. Natl Acad. Sci. USA. 2006;103:10901–10906. doi: 10.1073/pnas.0511156103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kondrashov DA, Zhang W, Roman Aranda, Stec B, Phillips GN., Jr Sampling of the native conformational ensemble of myoglobin via structures in different crystalline environments. Proteins. 2008;70:353–362. doi: 10.1002/prot.21499. [DOI] [PubMed] [Google Scholar]
- 23.Friedland GD, Lakomek NA, Griesinger C, Meiler J, Kortemme T. A correspondence between solution-state dynamics of an individual protein and the sequence and conformational diversity of its family. PLoS Comput. Biol. 2009;5:e1000393. doi: 10.1371/journal.pcbi.1000393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng J, McKnight CJ. Heterogeneity and dynamics in villin headpiece crystal structures. Acta Crystallogr. D Biol. Crystallogr. 2009;65:470–476. doi: 10.1107/S0907444909008646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Liu L, Koharudin LM, Gronenborn AM, Bahar I. A comparative analysis of the equilibrium dynamics of a designed protein inferred from NMR, X-ray, and computations. Proteins. 2009;77:927–939. doi: 10.1002/prot.22518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Prabhu NV, Lee AL, Wand AJ, Sharp KA. Dynamics and entropy of a calmodulin-peptide complex studied by NMR and molecular dynamics. Biochemistry. 2003;42:562–570. doi: 10.1021/bi026544q. [DOI] [PubMed] [Google Scholar]
- 27.Best RB, Clarke J, Karplus M. What contributions to protein side-chain dynamics are probed by NMR experiments? A molecular dynamics simulation analysis. J. Mol. Biol. 2005;349:185–203. doi: 10.1016/j.jmb.2005.03.001. [DOI] [PubMed] [Google Scholar]
- 28.Burra PV, Zhang Y, Godzik A, Stec B. Global distribution of conformational states derived from redundant models in the PDB points to non-uniqueness of the protein structure. Proc. Natl Acad. Sci. USA. 2009;106:10505–10510. doi: 10.1073/pnas.0812152106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Gutteridge A, Thornton J. Conformational changes observed in enzyme crystal structures upon substrate binding. J. Mol. Biol. 2005;346:21–28. doi: 10.1016/j.jmb.2004.11.013. [DOI] [PubMed] [Google Scholar]
- 32.Kumar S, Ma B, Tsai CJ, Sinha N, Nussinov R. Folding and binding cascades: dynamic landscapes and population shifts. Protein Sci. 2000;9:10–19. doi: 10.1110/ps.9.1.10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Goh CS, Milburn D, Gerstein M. Conformational changes associated with protein-protein interactions. Curr. Opin. Struct. Biol. 2004;14:104–109. doi: 10.1016/j.sbi.2004.01.005. [DOI] [PubMed] [Google Scholar]
- 34.Chothia C, Lesk AM. The relation between the divergence of sequence and structure in proteins. EMBO J. 1986;5:823–826. doi: 10.1002/j.1460-2075.1986.tb04288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bairoch A. The ENZYME database in 2000. Nucleic Acids Res. 2000;28:304–305. doi: 10.1093/nar/28.1.304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Porter CT, Bartlett GJ, Thornton JM. The Catalytic Site Atlas: a resource of catalytic sites and residues identified in enzymes using structural data. Nucleic Acids Res. 2004;32:D129–133. doi: 10.1093/nar/gkh028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Orengo CA, Pearl FM, Bray JE, Todd AE, Martin AC, Lo Conte L, Thornton JM. The CATH Database provides insights into protein structure/function relationships. Nucleic Acids Res. 1999;27:275–279. doi: 10.1093/nar/27.1.275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ortiz AR, Strauss CE, Olmea O. MAMMOTH (matching molecular models obtained from theory): an automated method for model comparison. Protein Sci. 2002;11:2606–2621. doi: 10.1110/ps.0215902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Gerstein M, Krebs W. A database of macromolecular motions. Nucleic Acids Res. 1998;26:4280–4290. doi: 10.1093/nar/26.18.4280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.van der Kamp MW, Schaeffer RD, Jonsson AL, Scouras AD, Simms AM, Toofanny RD, Benson NC, Anderson PC, Merkley ED, Rysavy S, et al. Dynameomics: a comprehensive database of protein dynamics. Structure. 2010;18:423–435. doi: 10.1016/j.str.2010.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]