Abstract
NCBI’s Conserved Domain Database (CDD) is a resource for the annotation of protein sequences with the location of conserved domain footprints, and functional sites inferred from these footprints. CDD includes manually curated domain models that make use of protein 3D structure to refine domain models and provide insights into sequence/structure/function relationships. Manually curated models are organized hierarchically if they describe domain families that are clearly related by common descent. As CDD also imports domain family models from a variety of external sources, it is a partially redundant collection. To simplify protein annotation, redundant models and models describing homologous families are clustered into superfamilies. By default, domain footprints are annotated with the corresponding superfamily designation, on top of which specific annotation may indicate high-confidence assignment of family membership. Pre-computed domain annotation is available for proteins in the Entrez/Protein dataset, and a novel interface, Batch CD-Search, allows the computation and download of annotation for large sets of protein queries. CDD can be accessed via http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml.
INTRODUCTION
The annotation of protein sequences with the location of domains is a common practice in the analysis of sequence data. The identification of a conserved domain footprint may be the only clue towards cellular or molecular function of a protein, as it indicates local or partial similarity to other proteins, some of which may have been characterized experimentally. Furthermore, the study of domain architectures in multi-domain protein families often reveals their evolutionary history and is a common tool in sequence classification. To this end, we released the first version of Conserved Domain Database (CDD) to the public in August 2000, >10 years ago, as a collection of 2738 multiple sequence alignment models, based on the content of the Pfam and SMART databases, and derived database search tools to support the rapid computation of sequence annotation. Since then, CDD has grown significantly both in volume and in scope. CDD now imports domain and protein family alignment models from Pfam (1) (currently mirroring version 24), SMART (2), COG (3), TIGRFAM (4) and the NCBI Protein Clusters database (5). It also contains a set of models curated by NCBI, many of which are organized into explicit hierarchies of homologous domain families that reflect functional divergence and divergent evolutionary processes. In addition, NCBI-curated domain models use 3D structure information explicitly, to define domain boundaries, guide multiple sequence alignment and provide insights into the relationship between sequence conservation and molecular function.
CDD is updated several times a year, with occasional updates initiated by new versions of imported data sets, and with most incremental updates reflecting additions to the NCBI-curated set of models. The current version of CDD, v2.25, contains 37 632 alignment models, of which 6056 have been curated by NCBI. Various aspects of CDD have been highlighted in earlier manuscripts (6); here we give a brief summary of major functionality pertaining to sequence annotation, some of which has been presented in greater detail in previous descriptions of CDD, and we introduce a novel tool, Batch CD-Search, that facilitates computation of annotation for large sets of protein queries.
SPECIFIC HITS, DOMAIN SUPERFAMILIES AND MULTI-DOMAIN MODELS
CDD is one of the many databases in NCBI’s Entrez query and retrieval system and can be searched, using the common Entrez interface, for keywords and terms indexed from names, titles and descriptions of the records. CDD is cross-linked with other databases such as Entrez Protein, PubMed and NCBI BioSystems, to name a few. However, most users of CDD encounter CDD records by following Conserved Domains links from Entrez/Protein sequence records, and also while executing protein BLAST and PSI-BLAST searches via NCBI’s web BLAST interface. The conserved domain model database can be scanned quickly with protein queries, and results showing domain annotation may already be available, while BLAST continues to scan the significantly larger non-redundant protein database. The application that visualizes live or pre-computed search results has been termed CD-Search (7), and the underlying algorithm is Reverse Position-Specific BLAST (RPS-BLAST), a variation of the commonly used PSI-BLAST method (8,9).
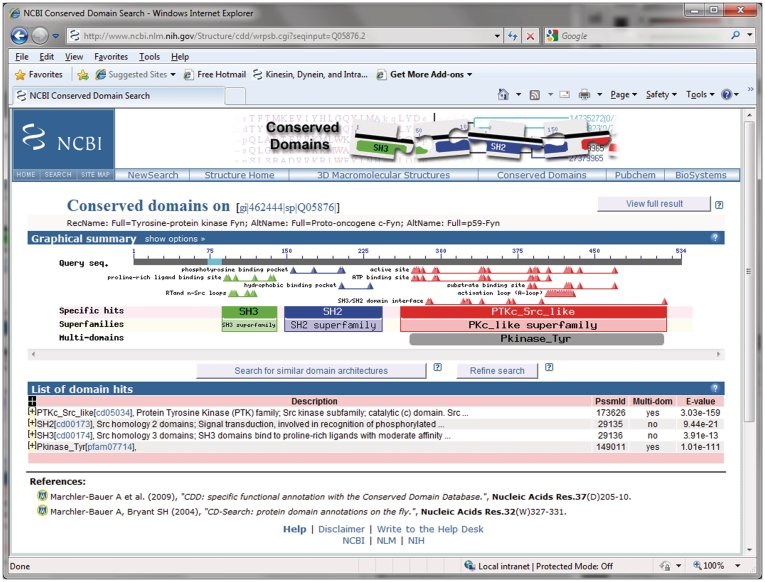
Figure 1 illustrates the layout of a page reporting conserved domain annotation. Live searches against the CDD will reproduce pre-computed search results unless the search parameters are modified from their default settings. Detailed descriptions of search result pages have been given previously (6). A concise domain annotation, as shown by default, will provide the locations of top-scoring domain footprints plus the locations of functional sites, which can be derived from the domain footprints. The locations are shown graphically, and detailed alignments are available as an option. Both CDD and CD-Search come with up-do-date help documentation that explains formatting and interpretation of output in detail, and which has been revised thoroughly in the past year. Domain footprints are shown as either:
Specific hits–indicating high confidence in the annotation with an NCBI-curated model, where the query model alignment score exceeds a model-specific threshold (10).
Superfamily annotation, where each superfamily is a collection of models representing homologous protein fragments, often quite redundant.
Annotation by multi-domain models, which have been excluded from the superfamily clustering as they tend to group non-homologous fragments into the same cluster.
Figure 1.
Conserved domain annotation on a well-characterized protein sequence. Shown here is the default concise view generated by the CD-Search tool, using pre-calculated alignment information. The view is divided into two panels: a graphical summary and a table detailing the individual matches. The query sequence coordinates are indicated on a gray bar in the top portion of the graphical summary. ‘Specific hits’ to NCBI-curated domain models are positioned in a separate area below the query sequence, with corresponding balloons rendered in saturated colors. The extent of the best-scoring hit for a region on the query also determines the annotation with the corresponding conserved domain ‘Superfamily’. ‘Superfamilies’ are positioned in the area below the ‘Specific hits’, and together these are enclosed in boxes to indicate superfamily membership of the NCBI-curated models. If the full (detailed) results display is selected, an area summarizing ‘Non-specific hits’ will be shown as well, and the corresponding boxes will be drawn so as to resolve their superfamily relationships; the highest ranked match for each superfamily defines the extents of the corresponding box. ‘Non-specific hits’ and ‘Superfamily’ balloons are rendered in pastel colors, with each superfamily being assigned a separate color. Matches to ‘multi-domain’ models are rendered as gray balloons in a separate area of the summary graph. Only the best-ranked non-overlapping multi-domain models are shown. Functional sites, as annotated on NCBI-curated domain models, are mapped to the query sequence and depicted as triangles. Sites are mapped from the highest ranked model only, and they are colored according to their source. Both conserved domain balloons and site annotations are hot-linked, so that moving the mouse over the objects displays additional information, and so that clicking on the objects launches conserved domain summary pages for the particular domain model, embedding the user query sequence in the alignment for further analysis, if applicable. A tabular view below the graphical summary lists E-values, multi-domain status and various identifiers for the conserved domain models identified as matches. The table rows can be expanded to display a detailed pair-wise sequence alignment between the query sequence and the domain model’s consensus sequence. An alignment of all sequences comprising a domain model, with or without the query sequence embedded, is accessible by clicking on the domain’s balloon representation in the graphical summary or its unique accession in the tabular summary, respectively.
By default, CD-Search displays only the highest ranking domain superfamily annotation for a given region on the query (and there can be no more than one specific hit, if any). The default display also shows only the highest ranked multi-domain model for a given query region, and only if that alignment is nearly complete with respect to the model. An alternative view shows the full alignment results, listing the individual models from all source databases that could be aligned to the query with significant scores. Often, the full alignment results are quite redundant.
FUNCTIONAL SITE ANNOTATION
Conserved Domain Models curated by NCBI often come together with the location and characterization of functional sites, such as active sites or binding sites for cofactors, nucleic acids, ions and polypeptides. These are recorded together with evidence, such as explicit complexes observed in experimentally determined 3D structure or the published literature. Sites are recorded only if it seems clear that they can be mapped onto a majority—if not all—members of the protein family modeled by the domain alignment. The query-to-model alignments computed by RPS-BLAST can be used to transfer site annotation onto the protein query. Currently, 13 562 sites have been recorded on 5214 models (∼86% of all NCBI-curated conserved domain alignments). Site annotation derived from CDD is visible in the default display of sequence records in the Entrez/Protein database, and functional site descriptions together with evidence can be examined in detail on the conserved domain summary pages, which are accessible via Entrez/CDD. The CDTree/Cn3D software, which is available for MS Windows and Mac OS X platforms, can be utilized to visualize conserved domain hierarchies, alignments, annotations, functional sites and corresponding evidence in great detail. CDTree and Cn3D are helper applications that can be launched via the conserved domain summary pages, and they are also the main curation tools used in the CDD project.
PROTEIN SEQUENCE ANNOTATION ON A LARGER SCALE
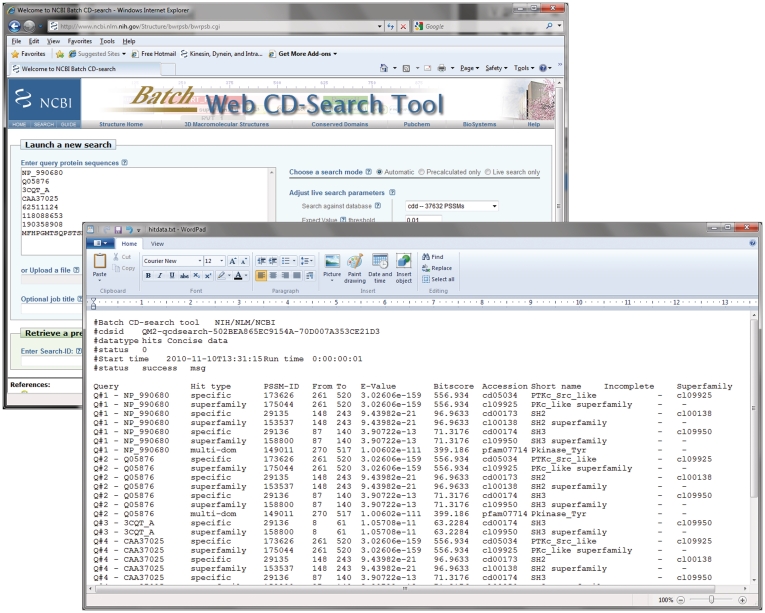
As pre-computed domain annotation is available for sequences in the Entrez/Protein database (excluding sequences associated with metagenomes), and as live searches for sequences not represented in Entrez can be run quickly, CDD may be used to compute and/or retrieve protein sequence annotation for large sets of query sequences. We have implemented a novel interface, Batch CD-Search, which facilitates the processing of upto 100 000 protein queries at a time. Queries can be supplied as either protein GIs (unique numerical identifiers used in Entrez/Protein), protein accessions or raw sequence data. Batch CD-Search then compiles the complete results, loading the domain hits on each query sequence into a temporary data base, which lets the user extract various results subsets, such as domain hits, alignment details and functional sites for up to several days after the search. The data can be downloaded in various formats including tab-delimited text (Figure 2), or displayed graphically within a web browser to show detailed annotations on any individual protein from the query list, using the ‘browse results’ function.
Figure 2.
The web-interface to Batch CD-Search. An input dialogue lets the user specify a set of protein queries or upload a corresponding file. The preliminary results page (not shown here) provides controls for downloading results in a variety of formats. The sample download format featured here lists one annotation per line, specifying the protein query, the type of domain hit (specific hit, superfamily or multidomain), from–to intervals on the query, E-value and score and the domain model’s name and accession. The Batch CD-Search help document describes the additional download options and formats available.
While large sets of queries can be uploaded conveniently via the web interface, Batch CD-Search can also be accessed programmatically via its URL; corresponding instructions are given in the help documentation.
Table 1 lists the Batch CD-Search URL, among other CDD-related resources. An alternative to using the Batch CD-Search service for the annotation of local data sets would be to run RPS-BLAST locally. CDD distributes pre-built search databases via the CDD FTP site, and also distributes individual position-specific score matrices (PSSMs), which can be subset arbitrarily, and/or combined with locally generated PSSMs to build special-purpose RPS-BLAST search databases.
Table 1.
URLs and other resources associated with the CDD project
| CDD | Database home page | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd.shtml |
| CDD help | CDD help documentation | http://www.ncbi.nlm.nih.gov/Structure/cdd/cdd_help.shtml |
| CDD FTP | CD models and alignments, pre-built search databases | ftp://ftp.ncbi.nih.gov/pub/mmdb/cdd |
| CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/cdd/wrpsb.cgi |
| Batch CD-Search | Live and pre-computed RPS-BLAST | http://www.ncbi.nlm.nih.gov/Structure/bwrpsb/bwrpsb.cgi |
| CDTree/Cn3D | Domain hierarchy viewer and editor | http://www.ncbi.nlm.nih.gov/Structure/cdtree/cdtree.shtml |
| rpsblast | Stand-alone tool for searching databases of profile models, part of the NCBI toolkit distribution |
|
FUNDING
Funding for open access charge: Intramural Research Program of the National Library of Medicine at the National Institutes of Health/DHHS.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank the authors of Pfam, SMART, COG, TIGRFAM and NCBI’s Protein Clusters database, and Paul Thiessen and the NCBI Information Engineering Branch for assistance with software development. They also thank the referees of the manuscript for helpful suggestions.
REFERENCES
- 1.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Letunic I, Copley RR, Pils B, Pinkert S, Schultz J, Bork P. SMART 5: domains in the context of genomes and networks. Nucleic Acids Res. 2006;34:D257–D260. doi: 10.1093/nar/gkj079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tatusov RL, Fedorova ND, Jackson JD, Jacobs AR, Kiryutin B, Koonin EV, Krylov DM, Mazumder R, Mekhedov SL, Nikolskaya AN, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Selengut JD, Haft DH, Davidsen T, Ganapathy A, Gwinn-Giglio M, Nelson WC, Richter AR, White O. TIGRFAMs and Genome Properties: tools for the assignment of molecular function and biological process in prokaryotic genomes. Nucleic Acids Res. 2007;35:D260–D264. doi: 10.1093/nar/gkl1043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Marchler-Bauer A, Anderson JB, Chitsaz F, Derbyshire MK, DeWeese-Scott C, Fong JH, Geer LY, Geer RC, Gonzales NR, Gwadz M, et al. CDD: specific functional annotation with the Conserved Domain Database. Nucleic Acids Res. 2009;37:D205–D210. doi: 10.1093/nar/gkn845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Marchler-Bauer A, Bryant SH. CD-Search: protein domain annotations on the fly. Nucleic Acids Res. 2004;32:W327–W331. doi: 10.1093/nar/gkh454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Marchler-Bauer A, Panchenko AR, Shoemaker BA, Thiessen PA, Geer LY, Bryant SH. CDD: a database of conserved domain alignments with links to domain three-dimensional structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fong JH, Marchler-Bauer A. Protein subfamily assignment using the conserved domain database. BMC Res. Notes. 2008;1:114. doi: 10.1186/1756-0500-1-114. [DOI] [PMC free article] [PubMed] [Google Scholar]