Abstract
Gene duplications are scattered widely throughout the human genome. A single-base difference located in nearly identical duplicated segments may be misjudged as a single nucleotide polymorphism (SNP) from individuals. This imperfection is undistinguishable in current genotyping methods. As the next-generation sequencing technologies become more popular for sequence-based association studies, numerous ambiguous SNPs are rapidly accumulated. Thus, analyzing duplication variations in the reference genome to assist in preventing false positive SNPs is imperative. We have identified >10% of human genes associated with duplicated gene loci (DGL). Through meticulous sequence alignments of DGL, we systematically designated 1 236 956 variations as duplicated gene nucleotide variants (DNVs). The DNV database (dbDNV) (http://goods.ibms.sinica.edu.tw/DNVs/) has been established to promote more accurate variation annotation. Aside from the flat file download, users can explore the gene-related duplications and the associated DNVs by DGL and DNV searches, respectively. In addition, the dbDNV contains 304 110 DNV-coupled SNPs. From DNV-coupled SNP search, users observe which SNP records are also variants among duplicates. This is useful while ∼58% of exonic SNPs in DGL are DNV-coupled. Because of high accumulation of ambiguous SNPs, we suggest that annotating SNPs with DNVs possibilities should improve association studies of these variants with human diseases.
INTRODUCTION
Gene duplication is considered to be a major contributor to genome evolution and consequent organism diversification. It can occur on the whole-genome scale, on blocks of genes or on single genes. Subjecting to evolutionary forces, duplicated genes become non-functionalized, subfunctionalized or neofunctionalized with the accumulation of mutations (1,2). Although mutations accumulated gradually, some recently duplicated regions may retain a high degree of sequence homology. Consequently, the genotyping data derived from these variations located in duplicated regions can resemble the genotyping results of single nucleotide polymorphisms (SNPs), as illustrated in Figure 1. A lack of detailed clarification of these undistinguishable SNPs located in duplicates can inflate the heterozygosity of SNPs. For instance, a number of annotated SNPs are associated with segmental duplication events (3). Many of these SNPs identified in duplicates may be nothing more than paralogous sequence variants (PSVs) that are defined as single nucleotide differences between duplicated loci in the genome and invariant in a population (4,5). Without supporting data of directly sequencing the lengthy targeted genomic region in numbers of individuals in the population, a SNP or a PSV in the highly conserved duplication often remains indistinguishable in genotyping. These ambiguous SNP calls are observed under current genotyping methods using assays of PCR, short-DNA sequencing and hybridization to DNA microarrays or beads. This confounding phenomenon is even more serious when identifying SNPs and mutations by shorter DNA fragments randomly produced from the next-generation sequencing (NGS). Because NGS data might contain a certain amount of short paralogous fragments, these paralogous variants may contribute to the false positive SNP/mutation calls in analysis. In addition, the released human reference genome has been frequently utilized as the foundation in genetic analysis since the Human Genome Project was completed in 2003. The existing duplication structure and paralogous variants in the reference genome may confound subsequent SNP detection. However, nucleotide variants in duplicated genes have not yet been mentioned and annotated, even though the concept of intergenic paralogous variations was previously discussed (6). Hence, we establish a genome-wide resource which is systematically generated from the reference genome to elucidate the ambiguity in human duplicated genes.
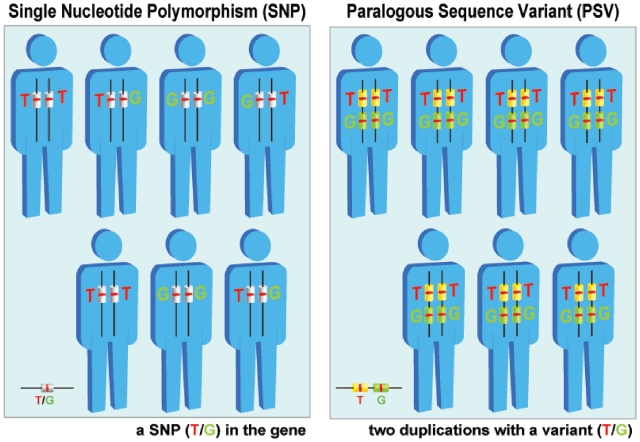
Figure 1.
SNPs versus Paralogous Sequence Variants (PSVs). The left panel displays a SNP (T/G) in the population. The base type of the specific genomic location varies among individuals as some are homozygous and some are heterozygous. The right panel illustrates PSVs. Two copies of the segment, duplications, exist in the genome. These two duplicates possess a different but invariant base (T/G) at the position of interest. The PSV may be undistinguishable from the SNP in genotyping.
As nearly identical DNA segments in the reference genome are the major interference of SNP calling, the duplication detection is crucial to the identification of duplicated gene nucleotide variants (DNVs). The published duplication studies mainly focus on large segmental duplications (7–9). For instance, the Human Paralogy Server comprises duplicates with a length of more than 1000 bases. However, controversial SNPs often were recognized as nucleotide variants surrounded by identical flanking sequences with the length shorter than few hundred bases. As a result, the existing duplication information is not suitable for the examination of SNP calls in duplicates. In addition, current high-throughput genotyping technologies can only handle short fragments. For example, the probe length of nucleotide SNP arrays is only tens. The sequence of NGS reads is shorter than 1000 bases. Furthermore, small-scale duplications occur through two major mechanisms. The first is the unequal crossover event that produces tandem duplications. Even though tandem duplications may diverge, they can still maintain sequence similarity through gene conversion. The other mechanism is retrotransposition, whereby the mRNA from a parental gene is converted into a cDNA fragment by reverse transcription and is then inserted into chromosomal DNA, forming an intronless paralog (10–12). Both kinds of gene-related duplicates are common in the human genome and preserve an extremely high-sequence similarity. Therefore, a proper duplication set for this issue is small scale and gene related. To perform a comprehensive examination and provide strictly qualified DNVs in our pipeline, we have applied a customized duplication detection method with extremely high-sequence similarities.
We first utilize reference transcripts as repeat units and apply a high degree of exonic sequence similarity to the single-gene duplication detection. The elimination of the interference from introns’ lower level of sequence conservation leads our identified duplicates to concentrate on functional regions and adapt to discover two mentioned duplicated gene types. One is that duplicated genes stemmed from segmental duplications, recent or functionally conserved duplicates, can be detected in our analysis. For example, NOMO1, NOMO2 and NOMO3 are tandem duplicates located on chromosome 16. Additionally, there is a duplicated cluster of CYP2D6 and its pseudogenes on chromosome 22q13.1. Both duplicated genes are all identified in our pipeline. The other is that retrogenes are naturally included in our analysis as they are intronless. For instance, we have identified POU5F1, which is located on chromosome 6 and possesses two retrotransposon insertion sites on chromosomes 8 and 12. This case demonstrates that our analysis can recognize intronless retrogenes. In sum, our algorithm can identify a comprehensive set of duplicated gene loci (DGL), which covers these two major types of gene-related duplicates.
The DNV database (dbDNV) collects nucleotide variants of DGL in the human reference genome via sequence alignments. Those DNVs could result in false increase of heterozygosity of SNP calls in genotyping. Referring to the dbDNV, researchers can focus on the DNV-coupled SNP calls to conduct in-depth investigations such as direct sequencing of longer genomic region. Therefore, they could exploit the valuable collected individuals to clarify those questionable observations while others usually are unable to do. This would also prevent researchers to report their detected SNPs in duplications with the PSV ambiguity. Public SNP records with the annotation of putative DNVs will increase the accuracy and reduce the subsequent interference in genetic association studies.
In the dbDNV, we have offered the browsing function and allowed different queries in the web interface. Users can examine DNVs located in the interested genes or genomic loci. The variation type, the multiple-sequence alignment of flanking sequences, and existing annotated SNP records in dbSNP are displayed in the web page. Aside from web exploration, users can download the flat file to perform their own batch analysis without any restriction. Currently, there is no existing resource systemically annotating paralogous variants in the human reference genome. We believe that dbDNV can promote more accurate and informative SNP/mutation annotations for duplicated genes.
DATA SOURCES AND IMPLEMENTATION
Data sources and tools
We used the human reference genome and reference transcripts from NCBI build 36.3 published on 26 March 2008 (http://www.ncbi.nlm.nih.gov/Genomes/). In this study, 37 312 reference transcripts (24 764 NMs and 12 548 XMs) were analyzed. To perform mRNA/DNA alignments, we downloaded the BLAST-Like Alignment Tool (BLAT) from the UCSC Genome Bioinformatics Site (http://genome.ucsc.edu/) (13). The SNP data set is downloaded from NCBI dbSNP build 130 released on 30 April 2009 (http://www.ncbi.nlm.nih.gov/projects/SNP/). The number of human reference SNP clusters (rs number) in build 130 is 17 804 034, and ∼99% of them (17 541 631) are mapped to the reference genome according to the b130_SNPContigLoc_36_3 table. The cross-reference information on human gene accession numbers in the dbDNV is obtained from Ensembl BioMart (http://www.ensembl.org/).
DGL assignment
We used BLAT to perform mRNA/DNA alignments for the human reference transcripts (Figure 2). BLAT on DNA is designed to quickly map sequences, whose lengths are >25 bases and whose similarities are >95%, without being affected by large insertions. Thus, we only considered BLAT results with highly aligned identity of transcripts as qualified alignments. On the basis of the BLAT results, 34 347 human reference transcripts possess their own unique genomic locations. The remaining transcripts (296,5) have multiple genomic locations with at least 95% identity in the sequence alignments. Transcripts associated with multiple locations may be transcribed from several genomic loci with a high degree of sequence similarity. We then designated these 2965 transcripts as duplicated genes and their corresponding genomic locations as DGL.
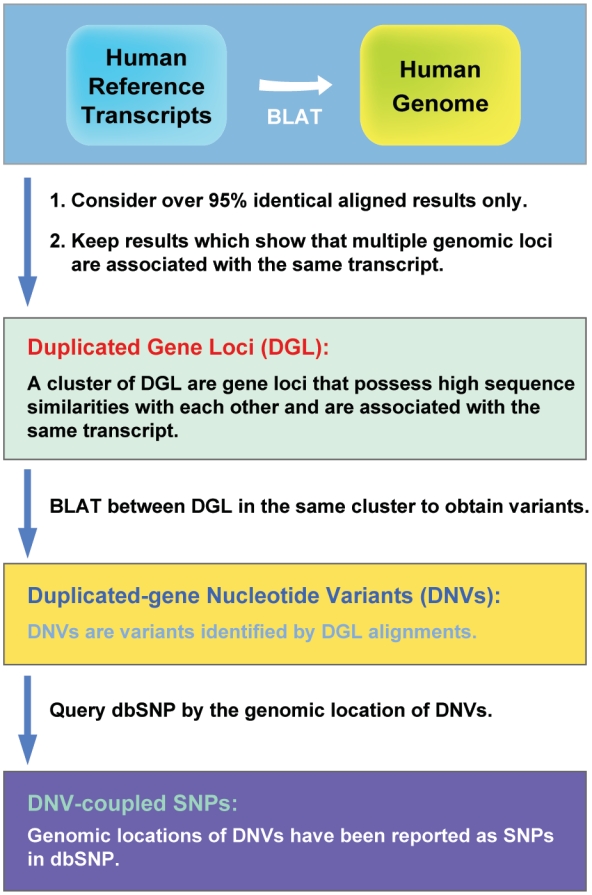
Figure 2.
Flowchart of the discovery pipeline. The flowchart describes the strategy used to identify DGL and their associated DNVs in human reference genome. The existing SNPs in dbSNP that occur at DNVs provide evidence of the ambiguity.
DNVs discovery
Our approach detects >10% of human genes, associated with 2965 reference transcripts, which are duplicated to possess multiple genomic loci. DGL cover two types of gene duplication events: segmental duplications and retrogenes. When gene loci are less homogeneous in introns or do not have introns, it is too difficult to carry out multiple sequence alignments. Here, we took advantage of BLAT, which can tolerate large insertions during the alignment of two duplicates. Two DGL can be aligned perfectly by BLAT, even if they possess diverged introns. Mismatches occurring in alignment are recorded as variants. The genomic positions of such variants in two aligned DGL are collected as DNVs. In total, we have identified 1 236 956 DNVs.
The DNV annotation
We have annotated DNVs by their flanking sequences and existing reference SNP records in dbSNP.
DNVs are nucleotide variants of DGL in the human reference genome. A cluster of DGL is genomic loci associated with the same transcript and possesses a high degree of the sequence similarity with each other. We first extracted the genomic sequence from the upstream 25 base to the downstream 25 base of each DNV. We then performed multiple sequence alignment on the extracted DNA segments. The flanking sequences of all corresponding DNVs were aligned together by ClustalW. The conservation of flanking sequences is displayed in the website.
We employed the b130_SNPContigLoc_36_3 table in dbSNP to retrieve the genomic location of reference SNPs. There are 523 896 reference SNPs with multiple genomic locations in dbSNP. About half of them locate in DGL. Further, there are 870 021 reference SNPs located in DGL. We queried dbSNP using the genomic locations of DNVs and obtained 304 110 reference SNPs (Figure 2). That is, one-third of reference SNPs within DGL resemble, identified DNVs. These SNPs are annotated as DNV-couple SNPs.
The maintenance and the future direction of dbDNV
The dbDNV is established to promote more accurate variation annotations on the basis of two major sources, the human genome assembly and SNP annotation from NCBI. The human genome is well-studied; therefore, the update frequency of the dbDNV would mainly depend on the data renewing of dbSNP. In addition, human genetic data from the 1000 genome project are rapidly accumulated. The dbDNV could incorporate the released data, especially pilot 3 data on gene regions, to reveal duplication structures and nucleotide variations of functional regions in the future.
DATABASE ACCESS
The main access to dbDNV is provided via the web interface at http://goods.ibms.sinica.edu.tw/DNVs. Three search functions have been established and described as following. Please refer to tutorial examples on search functions at http://goods.ibms.sinica.edu.tw/DNVs/search.html.
DGL
Users can obtain DGL by directly querying with the NCBI accession number of a reference transcript. Subjecting to a submitted transcript, the dbDNV provides genomic loci which are able to generate mRNA with >95% identity of the query. Some genomic loci may be already annotated with different gene names in the public database. This annotation is displayed in the ‘Annotated Gene’ column. Clicking this column leads users to the search page for identified DNVs within the selected gnomic locus. There is also a cross-reference search option for users to extract related RefSeq by different types of accession, e.g., gene symbol, Ensembl transcript ID or UniProt/SwissProt accession.
DNVs within a genomic region
Users can browse the identified DNVs in the genomic order by clicking the chromosome name. In addition to browsing by chromosomes, users can get a list of DNVs after specifying a chromosome or a genomic region. The dbDNV annotates each DNV by its 50-base flanking sequence. Users then can explore the related duplicating segments that generate the specified DNV by clicking the DNV’s position. The multiple sequence alignment of listed duplicating segments could demonstrate the interfering capability of the DNV via mimicking SNP calls in genotyping. Moreover, users can extend their search by click the DNV-related transcripts of the queried DNV. Furthermore, some DNVs may possess existing SNP records in dbSNP. If users click ‘rs_num’ instead of ‘position’, the dbDNV displays the genotype of the annotated SNP record accordingly. Users can also link out to dbSNP for detailed information.
DNV-coupled SNPs
This is the search page for looking at existing SNP records in dbSNP that are also identified as DNVs. Users can explore the DNV located on the same genomic position of the queried SNP. The related transcripts utilized to identify the DNV are listed. These transcripts are linked to the DGL search. Thus, users can use a preferred SNP record to pull down the comprehensive set of paralogous loci and their DNVs. In addition to the annotated genotype of the queried SNP, the variant’s flanking sequences in multiple duplicated loci are shown.
The first-search function is mainly employed to display the duplication structure of queried genes. The second one can provide DNV annotation within a user specified region. By manipulating these two search functions together, users explore the gene duplication structure and the associated DNVs. The dbDNV annotates DNVs with affiliated paralogous segments and existing SNP records in dbSNP. Users can perform in-depth investigation if necessary. This promotes more accurate SNP annotation. The third-search function is designed for examining existing SNP records with possibility of DNVs. As controversial SNP records have been accumulated in public, SNPs may require the inspection of existing in duplications before being further inferred. Aside from providing the search function, dbDNV also offers all users a flat file download without any restriction. Users can establish their own global genome examination system in genetic analysis on the basis of our DNV annotation.
RESULTS AND DISCUSSION
It requires a comprehensive set of short low-copy DNA sequences in paralogous genes to investigate DNVs that can mimic SNP calls. Because there is no database of short duplications available for this purpose, we have employed BLAT to identify DGL. In the BLAT results, the smallest length of aligned fragments is 25 bases, and sometimes this can be as small as 20 bases. Further, BLAT is designed to efficiently find DNA sequences whose similarities are >95% (13). According to the study performed by Nakken et al. (6), PSV-overlapping SNPs are inferred mainly from segmental duplications with 97–100% DNA sequence identity in intergenic regions. That is, providing short-aligned sequences with qualified sequence similarities makes BLAT suitable for our duplication identification. Aside from, controversial SNPs might be annotated with multiple genomic locations because they locate in nearly identical duplicates. We assumed that multilocated SNPs, SNPs annotated with multiple genomic locations in dbSNP, are unbiased distributed in human duplications. As a result, the coverage of multilocated SNPs is in proportion to the coverage of ambiguous SNPs. We employed this characteristic to inspect our identified DGL. Although DGL do not include intergenic duplications, they can still cover a significant amount of multilocated SNPs. Around half of all multilocated SNPs in dbSNP are presented in DGL; whereas, DGL comprise only 2.3% of the entire human genome. This high concentration of multilocated SNPs in DGL illustrates the efficiency of DGL coverage in our analytic pipeline. In addition, it is estimated that DGL cover a significant amount of ambiguous SNPs in dbSNP. More statistics are available on http://goods.ibms.sinica.edu.tw/DNVs/about.html.
We use transcripts as repeat units to locate the duplicated genomic loci instead of performing the indiscriminate base-to-base sequence alignment among chromosomes. It associates function and predicted gene structure with the identified duplicates. The identified duplications are no longer considered as repeat segments; rather, they can be regarded as genes undergoing evolutionary processes (14–17). A recent report showed that one out of five SNPs presented in intergenic duplicated regions bears a resemblance to PSVs (6). We show that over one-third of SNPs in DGL locate on the DNV positions and the percentage of exonic SNPs is up to 50%. The high concentration of controversial SNP records in DGL could be attributed by the evolutionary conservation of duplicated genes and resulted in the increase of genotyping difficulties to distinguish DNVs and those SNPs in duplicated genes. This highlights the importance of DNV annotation. The dbDNV can help genetic researchers clarify these ambiguities before the expiration of biological materials.
In addition to gene duplication, alternative splicing is another evolutionary mechanism that can increase functional variation by promoting gene diversification. After duplication, each duplicated locus has its own mutation accumulation. Once mutations hit the original splicing site, altered splicing patterns could lead the duplicated locus to evolve into a new gene. This evolving mechanism can be illustrated by the human gene NOMO. NOMO possesses three duplicated loci that have evolved into three different genes, termed NOMO1 (chr16:14835143-14897514), NOMO2 (chr16:18418683-18480929) and NOMO3 (chr16:16233958-16296168). There is a DNV (G/T) on the 5′-donor site of the thirty-first intron among these three duplicated loci (shown as an example of DNV-coupled search on http://goods.ibms.sinica.edu.tw/DNVs/search.html). As the 5′-GT donor site of the 31st intron was mutated to TT in chr16:14927643-14990014, this locus can only produce the form that retains the 31st intron. This demonstrates that each locus can develop its own expression pattern on the basis of a single mutation on splice sites. Issues of locus-specific transcripts and the differential expressed duplicating transcripts are of interest for gene dosage effects regarding to duplicated genes. This aspect can be a potential interesting application of the dbDNV.
Thus far, we have discussed the impact when variations occur in duplicates within a single genome. It is also essential to determine whether these duplicates exist constitutively in humans. If so, duplicated genes can demonstrate the hypothesis of functional compensation by close duplicates (18–20). Otherwise, a duplicate with different copy numbers across the population is so called a copy-number variation (CNV). This type of genetic variation represents a DNA segment that exhibits copy-number differences in the population (21–23). Hence, if some identified DNVs locate in CNVs, they may not be presented in every individual and increase the difficulty for using these DNVs in CNVs for genotyping analysis. Further, individual transcripts encoded from DNVs in CNVs owing to altered splicing sites can produce new type of genetic variations. These complex issues raise the demand for detailed DNV annotation in genetic studies.
FUNDING
Funding for open access charge: Academia Sinica, Taiwan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to acknowledge the valuable suggestions of Dr Ling-Hui Li and critical reading of the article by Dr Yuh-Shan Jou.
REFERENCES
- 1.Lynch M, Conery JS. The evolutionary fate and consequences of duplicate genes. Science. 2000;290:1151. doi: 10.1126/science.290.5494.1151. [DOI] [PubMed] [Google Scholar]
- 2.Ohno S. Evolution by gene duplication. Berlin, Germany: Springer; 1970. [Google Scholar]
- 3.Estivill X, Cheung J, Pujana MA, Nakabayashi K, Scherer SW, Tsui LC. Chromosomal regions containing high-density and ambiguously mapped putative single nucleotide polymorphisms (SNPs) correlate with segmental duplications in the human genome. Hum. Mol. Genet. 2002;11:1987–1995. doi: 10.1093/hmg/11.17.1987. [DOI] [PubMed] [Google Scholar]
- 4.Fredman D, White SJ, Potter S, Eichler EE, Den Dunnen JT, Brookes AJ. Complex SNP-related sequence variation in segmental genome duplications. Nat. Genet. 2004;36:861–866. doi: 10.1038/ng1401. [DOI] [PubMed] [Google Scholar]
- 5.Gut IG, Lathrop GM. Duplicating SNPs. Nat. Genet. 2004;36:789–790. doi: 10.1038/ng0804-789. [DOI] [PubMed] [Google Scholar]
- 6.Nakken S, Rodland EA, Rognes T, Hovig E. Large-scale inference of the point mutational spectrum in human segmental duplications. BMC Genomics. 2009;10:43. doi: 10.1186/1471-2164-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Li W-H, Gu Z, Cavalcanti A, Nekrutenko A. Detection of gene duplications and block duplications in eukaryotic genomes. J. Struct. Funct. Genom. 2003;3:27–34. [PubMed] [Google Scholar]
- 8.She X, Jiang Z, Clark RA, Liu G, Cheng Z, Tuzun E, Church DM, Sutton G, Halpern AL, Eichler EE. Shotgun sequence assembly and recent segmental duplications within the human genome. Nature. 2004;431:927–930. doi: 10.1038/nature03062. [DOI] [PubMed] [Google Scholar]
- 9.Durand D, Hoberman R. Diagnosing duplications–can it be done? Trends Genet. 2006;22:156–164. doi: 10.1016/j.tig.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 10.Brosius J. RNAs from all categories generate retrosequences that may be exapted as novel genes or regulatory elements. Gene. 1999;238:115–134. doi: 10.1016/s0378-1119(99)00227-9. [DOI] [PubMed] [Google Scholar]
- 11.Yu Z, Morais D, Ivanga M, Harrison P. Analysis of the role of retrotransposition in gene evolution in vertebrates. BMC Bioinformatics. 2007;8:308. doi: 10.1186/1471-2105-8-308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kaessmann H. More than just a copy. Science. 2009;325:958–959. doi: 10.1126/science.1178487. [DOI] [PubMed] [Google Scholar]
- 13.Kent WJ. BLAT–the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Suyama M, Harrington E, Bork P, Torrents D. Identification and analysis of genes and pseudogenes within duplicated regions in the human and mouse genomes. PLoS Comput. Biol. 2006;2:e76. doi: 10.1371/journal.pcbi.0020076. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Chauve C, Doyon J-P, El-Mabrouk N. Gene family evolution by duplication, speciation, and loss. J. Comput. Biol. 2008;15:1043–1062. doi: 10.1089/cmb.2008.0054. [DOI] [PubMed] [Google Scholar]
- 16.MacNeil AJ, McEachern LA, Pohajdak B. Gene duplication in early vertebrates results in tissue-specific subfunctionalized adaptor proteins: CASP and GRASP. J. Mol. Evol. 2008;67:168–178. doi: 10.1007/s00239-008-9136-y. [DOI] [PubMed] [Google Scholar]
- 17.Han MV, Demuth JP, McGrath CL, Casola C, Hahn MW. Adaptive evolution of young gene duplicates in mammals. Genome Res. 2009;19:859–867. doi: 10.1101/gr.085951.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Liang H, Li WH. Gene essentiality, gene duplicability and protein connectivity in human and mouse. Trends Genet. 2007;23:375–378. doi: 10.1016/j.tig.2007.04.005. [DOI] [PubMed] [Google Scholar]
- 19.Hsiao TL, Vitkup D. Role of duplicate genes in robustness against deleterious human mutations. PLoS Genet. 2008;4:e1000014. doi: 10.1371/journal.pgen.1000014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Qian W, Zhang J. Gene dosage and gene duplicability. Genetics. 2008;179:2319–2324. doi: 10.1534/genetics.108.090936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goidts V, Cooper D, Armengol L, Schempp W, Conroy J, Estivill X, Nowak N, Hameister H, Kehrer-Sawatzki H. Complex patterns of copy number variation at sites of segmental duplications: an important category of structural variation in the human genome. Human Genet. 2006;120:270–284. doi: 10.1007/s00439-006-0217-y. [DOI] [PubMed] [Google Scholar]
- 22.Lee S, Kasif S, Weng Z, Cantor CR. Quantitative analysis of single nucleotide polymorphisms within copy number variation. PLoS ONE. 2008;3:e3906. doi: 10.1371/journal.pone.0003906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sengupta M, Ray A, Chaki M, Maulik M, Ray K. SNPs in genes with copy number variation: a question of specificity. J. Genet. 2008;87:95–97. doi: 10.1007/s12041-008-0014-6. [DOI] [PubMed] [Google Scholar]