Abstract
Entrez Gene (http://www.ncbi.nlm.nih.gov/gene) is National Center for Biotechnology Information (NCBI)’s database for gene-specific information. Entrez Gene maintains records from genomes which have been completely sequenced, which have an active research community to submit gene-specific information, or which are scheduled for intense sequence analysis. The content represents the integration of curation and automated processing from NCBI’s Reference Sequence project (RefSeq), collaborating model organism databases, consortia such as Gene Ontology and other databases within NCBI. Records in Entrez Gene are assigned unique, stable and tracked integers as identifiers. The content (nomenclature, genomic location, gene products and their attributes, markers, phenotypes and links to citations, sequences, variation details, maps, expression, homologs, protein domains and external databases) is available via interactive browsing through NCBI’s Entrez system, via NCBI’s Entrez programming utilities (E-Utilities) and for bulk transfer by FTP.
INTRODUCTION
Entrez Gene is the gene-specific database at the National Center for Biotechnology Information (NCBI), a division of the National Library of Medicine, located on the campus of the US National Institutes of Health in Bethesda, MD, USA. Entrez Gene generates unique integers (GeneID) as stable identifiers for genes and other loci for a subset of model organisms. It tracks those identifiers and uses them to integrate multiple types of information including nomenclature, summary descriptions, accessions of gene-specific and gene product-specific sequences, chromosomal localization, reports of pathways and protein interactions, associated markers and phenotypes. Because the GeneID is used to represent gene-specific information in other databases at NCBI, the full Entrez Gene report includes a wealth of links to gene-specific literature citations, sequences, variations, homologs and databases outside of NCBI. Entrez Gene is integrated with NCBI’s Entrez system for interactive query, Linkout and access by E-Utilities (1).
Data in Entrez Gene result from integration of results from automated analyses and curation by Reference Sequence project (RefSeq) staff. Gene-specific annotation in sequences from NCBI’s RefSeq (2) or the International Nucleotide Sequence Database Collaboration (INSDC) (3) usually serves as the foundation, with value added by with information from collaborating model organism databases, public users and literature review (especially the Gene References into Function or GeneRIFs submitted by the public and staff of the National Library of Medicine). Updates are posted daily, and corrections or suggestions are welcomed (http://www.ncbi.nlm.nih.gov/RefSeq/update.cgi).
As of September 2010, there were almost 7 million current records in Entrez Gene, distributed among more than 7300 taxa (Table 1). Not all the taxa are represented comprehensively in Entrez Gene; most of the eukaryotes, for example, have records only for their mitochondrial or plastid genomes. The Gene Statistics site (http://www.ncbi.nlm.nih.gov/projects/Gene/gentrez_stats.cgi) reports both current and historical counts of records by taxonomic node and species. The history reports can be used to track the growth of the database. For example, the history of the eukaryotic node (http://www.ncbi.nlm.nih.gov/projects/Gene/gentrez_stats.cgi?HIS=1&TAXORG=2759) shows that from 2004 until the present the number of genes represented increased almost 10-fold (221997–2520683), with a 5-fold increase in the number of species (485–2265).
Table 1.
Representative statistics
| Category | Taxa | GeneIDs |
|---|---|---|
| Records with GO terms | 37 | 24 1633 |
| Records with GeneRIFs | 1475 | 59 627 |
| From archea/bacteria | 2290 | 4 090 330 |
| From fungi | 190 | 586 394 |
| From protozoa | 146 | 400 187 |
| From viruses | 2404 | 82 684 |
| From plants | 148 | 354 241 |
| From invertebrates | 670 | 554 008 |
| From vertebrates (non-mammalian) | 1090 | 134 355 |
| From mammalia | 311 | 471 725 |
FUNCTION OF THE DATABASE
A major goal of the database is to facilitate access to gene-specific information, and thus to expedite data exchange. The unique integer identifier assigned to each record (GeneID) is species specific. In other words, the integer assigned to dystrophin in human is different from that in any other species. The GeneID is reported in RefSeq records as a ‘db_xref’ (e.g. /db_xref= “GeneID:1756”, in GenBank format). The GeneID is also used to define genes in multiple files available for FTP, so that the information associated with GeneIDs is provided for unrestricted public use.
Entrez Gene is also key to representation of gene-specific information at NCBI. The information conveyed by establishing the relationship between sequence and a GeneID is used by many NCBI resources. For example, the names associated with GeneIDs are used in HomoloGene, UniGene and RefSeqs. The curated gene to sequence relationship reported in Entrez Gene is used to inform automated annotation of genomes and UniGene clustering.
WEB REPORTS
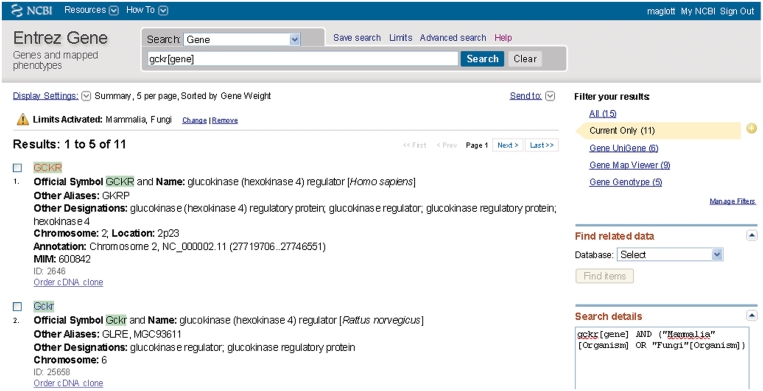
Entrez Gene provides multiple reports. For the interactive user, the defaults are web pages or files to download based on a query result, which are accessed by making selections revealed when ‘Display Settings’ or ‘Send to’ is activated (Figure 1).
The ‘summary display’ results from a query and provides the standard Entrez tools to navigate to information related to the set of records that matched the query (Figure 1).
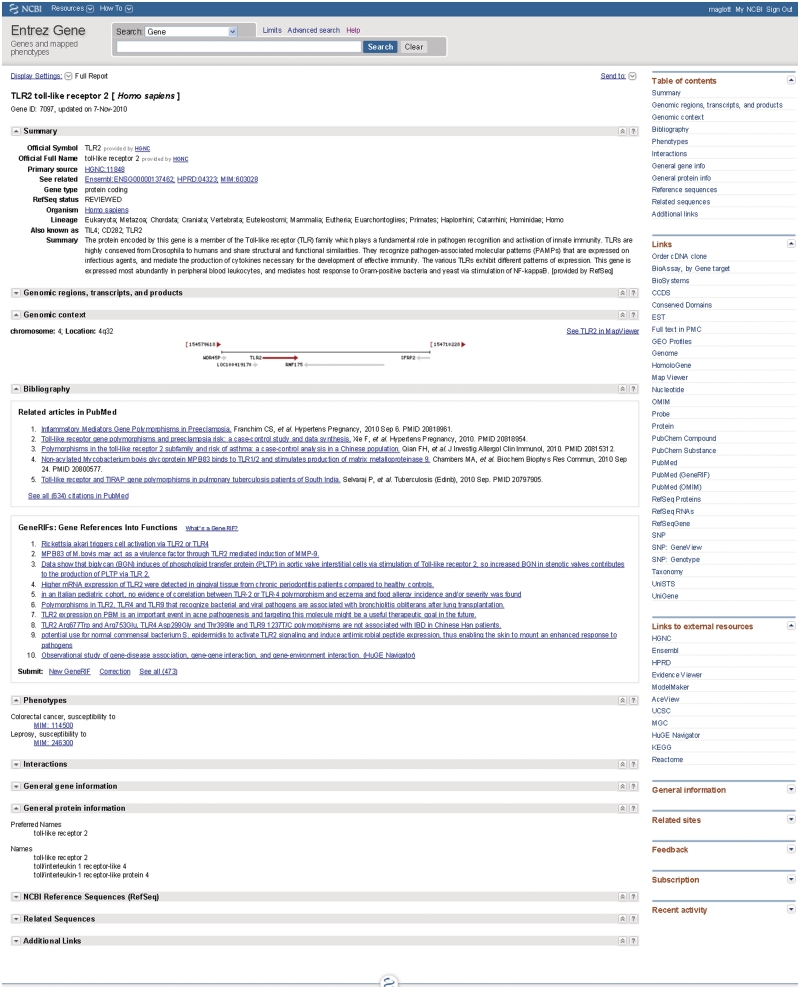
Each gene-specific ‘full report’ is accessed either by a gene-specific URL (e.g. http://www.ncbi.nlm.nih.gov/gene/7097) or by clicking on the symbol in the summary page (Figure 2).
The ‘Gene Table’ display (e.g. http://www.ncbi.nlm.nih.gov/gene/7097?report=gene_table) reports the intron/exon organization of the gene, as annotated on a RefSeq genomic sequence, with links to access the sequence of each exon, coding region or intron. If a gene is represented on multiple RefSeq genomic sequences, a menu is provided for the user to make a selection. The user can also elect to report the coordinates relative to the selected sequence or relative to the gene.
The ‘GeneRIF’ report (e.g. http://www.ncbi.nlm.nih.gov/gene/7097?report=GeneRif) provides a tabular display of GeneRIF texts, with the title and author(s) of each paper. Columns can be sorted by clicking on the column header.
The ‘XML’ and ‘ASN.1′ displays are provided as a text-like display without full Entrez functionality. If these pages are opened, the user must use the browser’s back function to return to the Entrez environment.
Text of the Summary, Full Report and GeneTable displays can be generated from the ‘Send to’ function at the top right, choosing File, and selecting an option from the menu.
Figure 1.
Representative ‘Summary’ report of query results. Result (partial) of a query to retrieve information about gckr as a gene symbol in mammals or fungi. This figure illustrates several points: (i) use of field restriction in the query; (ii) the display when ‘Limits’ is invoked to restrict results, in this case by species; (iii) use of ‘Display Settings’ to report five records per page ordered by Gene Weight (computed by number of gene-specific citations and conservation) and (iv) use of MyNCBI to highlight matches to the query term in the result set in green. ‘Limits Activated’: Mammalia, Fungi indicates that Mammalia and Fungi were both checked on the form accessed from ‘Limits’ over the query bar. Of the 15 results that were returned, the information under ‘Filter your results’ at the upper right indicate that 11 are current (Current Only, highlighted), 5 have genotype information available in dbSNP (Gene Genotype), 9 can be viewed in Map Viewer (Gene Map Viewer) and 8 have expression data in UniGene (Gene UniGene). For each GeneID returned by the filtered query, the summary includes the species, preferred and alternate symbols, preferred and other descriptive names, chromosome localization, the GeneID and the MIM number when appropriate. Click on any symbol to link to the full report or click on the Entrez Gene text at the upper left to return to Entrez Gene’s home page. The ‘Find related data’ menu in the column at the right allows selecting a database in which to find data related to initial query results. For example, to look for homologs of genes in a result set, select HomoloGene from the menu, read about how these links are calculated and click on ‘Find items’.
Figure 2.
Representative full report in Entrez Gene. This figure is based on http://www.ncbi.nlm.nih.gov/gene/7097 with several sections closed to allow the report to fit on one page. Note that the concepts enumerated in the Table of Contents at the upper right are provided explicitly in the Entrez Gene full report; concepts enumerated in the Links section are presented from other resources at NCBI. Some of the titles in the Links section do not correspond exactly to the name of an NCBI database. For example, RefSeq proteins result in a display in the Protein database; RefSeq RNA and RefSeqGene result in displays in the Nucleotide database and SNP GeneView results in the gene-specific display from dbSNP.
FTP AND E-UTILITIES
In addition to these views from Entrez, Gene provides a complete database extraction as well as several special reports for FTP transfer (ftp://ftp.ncbi.nlm.nih.gov/gene/README). Most of the files on the ftp site are refreshed daily. The data are also available from the programmatic interface to Entrez, namely E-Utilities (1).
CONTENT OF THE DATABASE
When are GeneIDs assigned and how is each categorized?
A GeneID is usually assigned to what is annotated as a gene on a RefSeq record. Exceptions include RefSeqs from bacterial genomes that are annotated whole-genome shotgun sequences. A GeneID may also be assigned when no RefSeq exists. This may occur when an authoritative source for a genome, such as a model organism-specific database, assigns an identifier to what is termed a gene, mapped locus or trait, even though that entity is not completely defined by sequence. When a record in Entrez Gene is established, it is assigned a category (e.g. protein coding, pseudogene, rRNA, unknown) consistent with the molecule types defined by the INSDC. The term ‘unknown’ is used when the category is under review by RefSeq staff, as when some of the sequences defining the gene are annotated with coding regions, but the support for that annotation is inconclusive. The category can change without changing the GeneID.
A representative full record
A full record in Entrez Gene is subdivided into content-specific sections as summarized in its table of contents and the section headers (Figure 2). Each section of the record can be collapsed, and the section divider has both a link (icon: question mark) to documentation and function to return to the top of the page. Not all records will have content in each category, but all have a GeneID, names and information supporting the creation of the record (either sequence, link to an external database or publications). Some of the content is not reviewed by NCBI staff, but integrated automatically. For example, the content in the Interactions section, and several sections of the General Gene Information sections are primarily from external groups [e.g. EcoCyc (4), Gene Ontology Consortium (5), KEGG (6), Reactome (7)]. When genomic RefSeqs annotated with the gene are available, the ‘Genomic regions, transcripts and products’ section includes an embedded, interactive sequence display that can be expanded. To expedite loading of web pages, the default display of the full record often renders only a subset of the bibliographic and interaction information. Links are provided within those sections to navigate to additional pages. To get the full report in one page, the ‘Send to’ option allows saving the record as a text file.
Comprehensive and up-to-date documentation of the contents and maintenance of these sections are provided in the Gene Help Book on NCBI’s bookshelf (http://www.ncbi.nlm.nih.gov/books/NBK3839/).
In addition to the content it displays directly, Entrez Gene provides numerous links to information from other databases within the text and in the Links menu at the right (Figure 2). For example, clicking on ‘RefSeq protein’, ‘RefSeq RNA’ or RefSeqGene in the menu at the right takes users to the Nucleotide database where the RefSeq records specific to one gene can be retrieved, reviewed and analyzed. Similarly, users may select HomoloGene or ProteinClusters (8) links for integration of information about homologs, Map Viewer for extended genomic context and comparative maps, GENSAT, UniGene and GEO for expression data, Conserved Domain Database for domain content of proteins, OMIM (9) for human Mendelian disorders, PubMed and Books for publications. Entrez Gene also provides extensive links to species- or gene-specific databases or gene records in other browsers. Many groups also use the LinkOut (1) method to link their resources to information in Entrez Gene. The integration of explicit content links to gene-specific reports in other NCBI databases, and links to external resources all contribute to making Entrez Gene an effective site to retrieve gene-specific information.
ACCESS TO ENTREZ GENE
The information in Entrez Gene can be accessed in multiple ways at NCBI (Table 2). The simplest way is to submit an interactive query to Entrez from the NCBI home page and display the results in Gene, or enter a query in any Entrez query bar and restrict the database search to Gene. Starting from Entrez Gene directly, the ‘Limits’ and ‘Advanced Search’ pages make it easier to construct complex queries and submit them. For example, the ‘Limits’ page supports finding genes by chromosome location or in a taxonomic node and the ‘Advanced Search’ page has a query builder, a function to browse all the terms in the database and the fields in which they occur (browse index) and a tool to combine and compare previous query results (search history). All the text in the Entrez Gene record is indexed to support retrieval. For a more comprehensive discussion on how to query Entrez Gene, please refer to the Query Tips section of the help documentation. If the location in the record that matches a query term is not immediately obvious, the text of interest may be in the next page of a paginated section.
Table 2.
Accessing Entrez gene
| Direct query | |
|---|---|
| Enter search term(s) and select results shown in the Gene section | http://www.ncbi.nlm.nih.gov or http://www.ncbi.nlm.nih.gov/gquery/gquery.fcgi? |
| Enter search term(s) and query only Entrez gene | http://www.ncbi.nlm.nih.gov/gene or select Gene as the search option from any Entrez query bar |
| E-Utilities: check the result interactively. (Hint: view source if the browser does not display the XML.) | http://eutils.ncbi.nlm.nih.gov/entrez/eutils/esummary.fcgi?db=gene&id =672&retmode=xml l |
| Record-specific connections to gene from other NCBI databases | |
| Gene option in the links menu located at the right of a display | Click on gene to find gene records related to the record being displayed. |
| Gene ‘ads’ | A query that looks like a gene symbol results in a gene Ad (located above the query results) suggesting users to check Entrez Gene for additional information; or, for sequence records with explicit links, an Ad is provided in the right column to highlight the link to Entrez gene. |
| Gene symbols or GeneID | Many NCBI databases provide links to Entrez Gene anchored on either the gene symbol or the GeneID. |
| Links called Gene or G | Map Viewer’s annotation of Genes; BLAST retrieval of accessions connected to Gene records. |
| More information | |
| Help documentation | http://www.ncbi.nlm.nih.gov/books/NBK3839/ |
| General use of Entrez | http://www.ncbi.nlm.nih.gov/books/NBK3836/ |
Another way to access Entrez Gene is to take advantage of links computed by the Entrez system (1). For example, users starting at PubMed may use the ‘Find related data’ or ‘All links from this record’ options to discover records in Entrez Gene connected to the publication(s). The BLAST group uses the GeneID–sequence relationship maintained by Entrez Gene to help users navigate from protein or mRNA accessions matching a sequence query to Entrez Gene via the blue G icon. Map Viewer provides links from annotated genes to Entrez Gene. And RefSeq records include the GeneID as a db_xref in the gene feature. Thus, users can navigate to Entrez Gene not only by text but also by genomic position, RefSeq annotation and sequence data (BLAST, Nucleotide, Protein).
Users are encouraged to register for MyNCBI (http://www.ncbi.nlm.nih.gov/books/NBK3843/). which supports registering searches and receiving e-mails when records are created or updated. It also supports customizing the display to identify what subset of records returned by a query has particular attributes.
FUTURE DIRECTIONS
The number of records in Entrez Gene will continue to increase as new species are sequenced and genes are identified. During 2011, sections will be added to the web interface and/or the content will be enhanced so that users will be provided more information in the full report before navigating to related sites at NCBI. This transition was started in 2010 with the addition of the phenotype section. Finally, as new databases with gene-specific content are implemented at NCBI, content and/or links will be added to Entrez Gene.
FEEDBACK
We welcome feedback with respect to the Entrez Gene interface or any data contained therein. Please select from the Feedback options on any Gene page (Figure 1).
FUNDING
Funding for open access charge: The Intramural Research Program of the National Institutes of Health; National Library of Medicine.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Pruitt KD, Tatusova T, Klimke W, Maglott D. NCBI Reference Sequences: current status, policy and new initiatives. Nucleic Acids Res. 2009;37:D32–D36. doi: 10.1093/nar/gkn721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Keseler IM, Bonavides-Martínez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37:464–470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ogata H, Goto S, Sato K, Fujibuchi W, Bono H, Kanehisa M. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 1999;27:29–34. doi: 10.1093/nar/27.1.29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2010;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Klimke W, Agarwala R, Badretdin A, Chetvernin S, Ciufo S, Fedorov B, Kiryutin B, O’Neill K, Resch W, Resenchuk S, et al. The National Center for Biotechnology Information’s Protein Clusters Database. Nucleic Acids Res. 37:D216–D223. doi: 10.1093/nar/gkn734. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Onliine Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]