Abstract
MicroRNAs (miRNAs), one type of small RNAs (sRNAs) in plants, play an essential role in gene regulation. Several miRNA databases were established; however, successively generated new datasets need to be collected, organized and analyzed. To this end, we have constructed a plant miRNA knowledge base (PmiRKB) that provides four major functional modules. In the ‘SNP’ module, single nucleotide polymorphism (SNP) data of seven Arabidopsis (Arabidopsis thaliana) accessions and 21 rice (Oryza sativa) subspecies were collected to inspect the SNPs within pre-miRNAs (precursor microRNAs) and miRNA—target RNA duplexes. Depending on their locations, SNPs can affect the secondary structures of pre-miRNAs, or interactions between miRNAs and their targets. A second module, ‘Pri-miR’, can be used to investigate the tissue-specific, transcriptional contexts of pre- and pri-miRNAs (primary microRNAs), based on massively parallel signature sequencing data. The third module, ‘MiR–Tar’, was designed to validate thousands of miRNA—target pairs by using parallel analysis of RNA end (PARE) data. Correspondingly, the fourth module, ‘Self-reg’, also used PARE data to investigate the metabolism of miRNA precursors, including precursor processing and miRNA- or miRNA*-mediated self-regulation effects on their host precursors. PmiRKB can be freely accessed at http://bis.zju.edu.cn/pmirkb/.
INTRODUCTION
The biological significance of miRNAs was widely recognized at the beginning of this century. Since then, miRNAs have been extensively studied in both plants and animals (1,2). The miRNAs, ∼21 nt in length, can recognize their targets based on complementary recognition sites, normally resulting in transcript cleavages in plants or translational repression in animals (1,2). The functionalities of miRNA targets vary widely, although a sizable portion were transcription factors (2,3), suggesting extraordinarily complex regulatory networks that miRNAs involved in. To date, many miRNA families, either highly conserved or species-specific, have been cloned and functionally characterized (2,3). With the advent of next-generation sequencing, significant advances have been achieved in the miRNA research area, and valuable sequencing data were generated (4,5). Thus, standardized databases are required for data deposition, organization, parsing and analysis, while also allowing for user queries. Currently, there are several established miRNA databases, such as miRBase (6) and the plant microRNA database (PMRD) (7). These two databases catalog various organisms, and numerous plant species, respectively. Although extremely comprehensive, they only provide general information for miRNAs, such as sequences, secondary structures, experimental evidences, and references. Hence, by using such kind of databases, only basic queries of specific miRNAs are available. Apparently, a need exists for databases providing more specific tools and in-depth information regarding the miRNAs.
Here, we focus on the miRNAs of two model plants, the eudicot Arabidopsis and the monocot rice. PmiRKB (http://bis.zju.edu.cn/pmirkb/) contains four major functional modules which are titled: ‘SNP’, ‘Pri-miR’, ‘MiR–Tar’ and ‘Self-reg’ (Figure 1). The ‘SNP’ module contains SNPs within the pre-miRNAs among different Arabidopsis or rice subspecies based on currently released SNP data. To investigate the potential effects of SNPs on the secondary structure transformation of pre-miRNAs, the stem–loop structured pre-miRNAs were predicted. The ‘SNP’ module also includes SNPs present in the mature miRNAs and their target sites, which may affect miRNA–target interactions. The second module, ‘Pri-miR’, provides data regarding the transcriptional contexts of pre- and pri-miRNAs, which include tissue-specific considerations. To our knowledge, this is the first large-scale attempt to elucidate the transcriptional ranges of pri-miRNAs in planta. The third module, ‘MiR–Tar’, can identify cleavage signals present within predicted miRNA–target recognition sites based on PARE data derived from plant degradomes. As a result, this module can serve as the reference for in vivo miRNA–target pair validation. Lastly, the ‘Self-reg’ module provides data regarding miRNA precursor processing, and miRNA- or miRNA*-mediated cleavage effects on their host precursors, based on cleavage signals detected by PARE. Together, our online service of PmiRKB implemented in PostgreSQL + Apache + hypertext preprocessor (PHP) + scalable vector graphics (SVG) provides an unprecedented resource for plant miRNA research, ensuring its value for biologists.
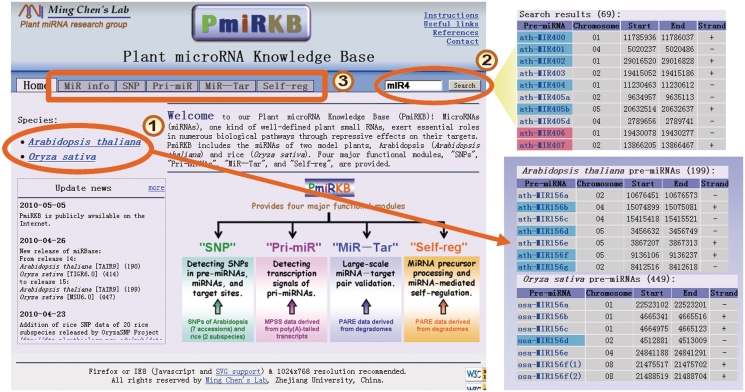
Figure 1.
Overview of the accesses to the functional modules of PmiRKB. There are three options to perform queries of the data maintained in PmiRKB. In this figure they are circled or boxed and are numbered 1 to 3. The first is an option to search Arabidopsis thaliana or Oryza sativa miRNAs. An example of each species’ output format is shown in the purple shaded box on the lower right of the figure as indicated by the arrow. Second, there is an option to perform a search of the entire knowledge base. An example of the output for the search query is shown on the upper right of the figure to the right of the yellow triangle. Third, there are five functional modules: ‘MiR info’, ‘SNP’, ‘Pri-miR’, ‘MiR–Tar’ and ‘Self-reg’, which can be clicked on to access the various datasets described in the illustrated boxes below. Note that in both the miRNA lists and the search results (shown on the right panels), the pre-miRNAs with SNPs in their mature miRNAs are shaded in red, and those with SNPs in other regions of the pre-miRNAs are shaded in blue. Additional resources are also available that include (clockwise from the top right on the PmiRKB homepage): ‘Instructions’, ‘Useful links’, ‘References’, ‘Contact’, an illustrated overview of the four major modules of PmiRKB, as well as a panel that provides ‘Update News’.
DATA COLLECTION
Sequence information for the miRNAs included in PmiRKB was retrieved from miRBase (http://www.mirbase.org/index.shtml; release 15) (6). Genomic information for Arabidopsis and rice were downloaded from the Arabidopsis information resource (TAIR release 9, ftp://ftp.arabidopsis.org/home/tair/Sequences/) (8), and the rice genome annotation project established by the institute for genome research (currently named the J. Craig Venter Institute) (TIGR rice genome release 6.1, ftp://ftp.plantbiology.msu.edu/pub/data/Eukaryotic_Projects/o_sativa/annotation_dbs/) (9), respectively. The SNPs of seven Arabidopsis accessions (Col-0, Bur-0, Tsu-1, Cvi-0, Ler-1, Bay-0, and Sha; the genome of Col-0 served as the reference) reported by Weigel’s group (10) were retrieved from TAIR (ftp://ftp.arabidopsis.org/home/tair/Genes/TAIR9_genome_release/TAIR9_gff3/Variation_GFF/, release 9) (8). The SNPs between two rice subspecies, Nipponbare and 93–11 (the genome of Nipponbare served as the reference), were a gift from the Paterson group (11), while the SNPs belonging to 20 rice subspecies (Nipponbare, Tainung 67, Li-Jiang-Xin-Tuan-Hei-Gu, M 202, Azucena, Moroberekan, Cypress, Dom-Sufid, N 22, Dular, FR13 A, Aswina, Rayada, IR64-21, Shan-Huang Zhan-2, Pokkali, Swarna, Sadu-Cho, Minghui 63 and Zhenshan 97B; the genome of Nipponbare served as the reference) were obtained from the OryzaSNP Project (ftp://ftp.plantbiology.msu.edu/pub/data/Oryza_SNP/) (12,13). All of the SNPs, and their genomic positions, were manually checked based on the current genomic information available from TAIR (release 9) (8) and TIGR (release 6.1) (9). Detected inconsistencies were removed.
A majority of the MPSS and the PARE data of Arabidopsis and rice were retrieved from the plant MPSS databases (http://mpss.udel.edu/at/mpss_index.php and http://mpss.udel.edu/at_pare/ for Arabidopsis, and http://mpss.udel.edu/rice/mpss_index.php and http://mpss.udel.edu/rice_pare/ for rice, respectively) (14–16). In addition, a portion of PARE data of Arabidopsis and short-read sequences derived from poly(A)-tailed transcripts and degradomes in rice were collected from recent reports (17–19). For users, more detailed descriptions of the features and the usages of these high-throughput sequencing (HTS) data can be obtained from the link ‘Instructions’ of PmiRKB.
All the data retrieved for this study were summarized in Table 1.
Table 1.
Data sources used for the construction of PmiRKB
MicroRNA TARGET PREDICTION
MiRNA targets were predicted in silico using the algorithm employed by miRU with default settings (20), and the messenger RNA (mRNA) sequences of Arabidopsis and rice were retrieved from TAIR (release 9) and TIGR (release 6.1), respectively. A random subset of the predicted results was manually checked with the results generated by miRU to ensure the accuracy of our prediction program written in C language.
DATABASE CONSTRUCTION AND CONTENT
SNPs present in pre-miRNAs and mature miRNAs in Arabidopsis and rice were extracted from the whole-genome SNP datasets, and downstream targets were predicted for each miRNA using the algorithm employed by miRU (20). These results were integrated into PmiRKB, along with the SNPs present in predicted miRNA target sites and the SNPs with potential to alter the secondary structures of pre-miRNAs according to RNAfold prediction (21) (see Table 2 for summary). The resulting ‘SNP’ module can identify different secondary conformations of a specific pre-miRNA among various Arabidopsis or rice subspecies, which result from SNP-induced thermal stability alteration (Figure 2A). Moreover, SNPs present in the miRNA–target binding regions, which have the potential to directly disturb miRNA–target interactions, were taken into account (Figure 2A). Accordingly, the output from the ‘SNP’ module can direct the investigation of phenotypic or physiological divergences among different Arabidopsis or rice subspecies, which may be attributed to SNP-involved, miRNA-mediated regulatory processes.
Table 2.
Statistical result of SNPs used for the construction of PmiRKB
| Species | Subspecies | Pre-miRNAsa | MiRNAsb | Target sitesc | Othersd | Sume |
|---|---|---|---|---|---|---|
| Arabidopsis | Bur-0 | 148 | 11 | 239 | 569 414 | 569 795 |
| Tsu-1 | 147 | 11 | 218 | 500 987 | 501 349 | |
| Cvi-0 | 213 | 23 | 343 | 807 540 | 808 085 | |
| Ler-1 | 66 | 3 | 145 | 304 370 | 304 580 | |
| Bay-0 | 79 | 12 | 119 | 316 805 | 316 999 | |
| Sha | 105 | 12 | 156 | 402 326 | 402 580 | |
| Sumf | 758 | 72 | 1220 | 2 901 442 | 2 903 388 | |
| Rice | Tainung 67 | 0 | 0 | 0 | 228 | 228 |
| Minghui 63 | 9 | 0 | 17 | 60 681 | 60 707 | |
| Zhenshan 97B | 7 | 0 | 12 | 38 653 | 38 672 | |
| Sadu-Cho | 10 | 0 | 15 | 60 634 | 60 659 | |
| N 22 | 4 | 0 | 25 | 58 266 | 58 295 | |
| FR13 A | 6 | 1 | 24 | 66 304 | 66 334 | |
| Moroberekan | 0 | 0 | 8 | 22 098 | 22 106 | |
| Dom-Sufid | 0 | 0 | 1 | 2185 | 2186 | |
| Aswina | 6 | 0 | 18 | 66 834 | 66 858 | |
| Dular | 9 | 1 | 30 | 63 690 | 63 729 | |
| Rayada | 9 | 1 | 29 | 66 053 | 66 091 | |
| Pokkali | 8 | 0 | 18 | 55 738 | 55 764 | |
| IR64-21 | 6 | 0 | 15 | 58 244 | 58 265 | |
| Azucena | 2 | 0 | 4 | 15 348 | 15 354 | |
| Shan-Huang Zhan-2 | 7 | 0 | 13 | 48 100 | 48 120 | |
| Li-Jiang-Xin-Tuan-Hei-Gu | 0 | 0 | 1 | 3206 | 3207 | |
| Swarna | 11 | 1 | 16 | 54 020 | 54 047 | |
| Cypress | 2 | 1 | 7 | 23 959 | 23 968 | |
| M 202 | 0 | 0 | 0 | 5493 | 5493 | |
| 93-11 | 48 | 6 | 89 | 382 279 | 382 416 | |
| Sumf | 144 | 11 | 342 | 1 152 013 | 1 152 499 |
aNumber of SNPs that reside in pre-miRNAs (precursor microRNAs) in each subspecies.
bNumber of SNPs that reside in mature microRNAs in each subspecies.
cNumber of SNPs that reside in the microRNA target recognition sites in each subspecies.
dNumber of SNPs with other genomic locations in each subspecies.
eTotal number of SNPs in each subspecies.
fSum of the SNPs in all the subspecies belonging to each categories.
Note that, the SNPs within pre-miRNAs, mature microRNAs and microRNA target recognition sites may overlap with each other, resulting in the inconsistencies between ‘Sume’ and the sum of all the categories (‘Pre-miRNAsa’, ‘MiRNAsb’, ‘Target sitesc’ and ‘Othersd’) in each subspecies.
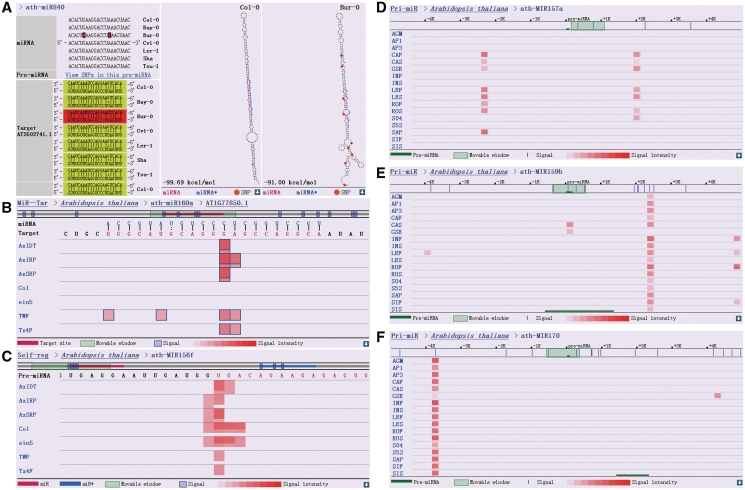
Figure 2.
Output data from the four major modules of PmiRKB. (A) An example of SNPs identified in the ‘SNP’ module (top left panel, highlighted in dark pink), which have the potential to influence the stability of miRNA–target RNA duplexes (lower left panel, indicated by different background colors). The effect of SNPs (indicated by orange circles) on the secondary structure of a specific pre-miRNA (precursor miRNA), ath-MIR840 for example, is illustrated on the right two panels. (B) Output data from the ‘MiR–Tar’ module that indicates AT1G77850.1 is a genuine target of ath-miR160a, based on the intense PARE signals detected in the middle of the miRNA–target duplex. The 100-nt sequence (gray horizontal line) surrounding the target site (red horizontal line) is shown. The blue outlines of the PARE signals denote the signatures that possess unique loci in the corresponding transcriptome. (C) The result given by ‘Self-reg’ indicates that the Dicer-like 1 (DCL1)-mediated first-step cleavages on the stem region of a specific pri-miRNA (primary miRNA) can be reflected by PARE data. In most cases, cleavage signals generated from the left arms of the pri-miRNAs were much easier to be detected than those from the right arms, likely due to the longer remnants generated from the left ones after DCL1-mediated cleavages. Cleavage signals within the miRNA* region of the pre-miRNA, ath-MIR156f, may represent miRNA-mediated self-regulation. For both (B and C), the signal intensity at a specific position is enhanced by the supporting of distinct signatures (represented by the red overlapping signals in different lengths in vertical orientation). (D, E and F) are output data from the ‘Pri-miR’ module. (D) represents tissue-specific transcription signals associated with the 3′-downstream region of ath-MIR157a, which could be hardly observed in the reproductive organs including inflorescences and siliques (the libraries lack of related signals, i.e. AGM, AP1, AP3, INF, INS, SIF, and SIS, were prepared from the reproductive organs of Arabidopsis). The transcription signals with distinct tissue-origin patterns were observed surrounding certain pre-miRNAs, such as ath-MIR159b (E) and ath-MIR170 (F), indicating that independent transcription units may exist. For (D, E and F), the signal intensity at a specific position is enhanced by the supporting of distinct signatures (represented by the red overlapping signals in different lengths in horizontal orientation).
HTS data, largely represented by MPSS data (22), were retrieved from public resources (see details in ‘Data Collection’ section). These short reads were derived from sequencing of the poly(A)-tailed transcripts in various tissues (14). Although many poly(A)-tailed transcripts are represented by mRNAs, a majority of pri-miRNAs transcribed by RNA polymerase II also possess poly(A) tails (2,23). Thus, these HTS datasets can be used to detect the transcriptional signals of pri-miRNAs in a tissue-specific manner. For this purpose, all the HTS signatures were mapped to the corresponding genomes, and only the signatures with unique genomic loci were reserved for further investigation. The short reads located within ∼5 kb on either side of a pre-miRNA were used to reveal the transcriptional context of the corresponding pri-miRNA. Moreover, expression levels of the short reads associated with each library prepared from distinct tissues were normalized in RPM (reads per million) to allow cross-library comparisons. Thus, the transcriptional signals of certain pri-miRNA can be queried in a tissue-specific manner by using the module ‘Pri-miR’ (Figure 2D–F).
In plants, most miRNAs cleave their downstream targets depending on the highly complementary recognition sites (2). The resulting 3′ cleavage remnants are usually utilized for miRNA–target pair validation and slicing site mapping by using modified 5′ rapid amplification of cDNA ends (RACE) (3). However, this classical method would be time-consuming, tedious, and costly once applied to large-scale validation of miRNA–target pairs. To overcome these limitations, the modified 5′ RACE has been combined with the newly available HTS to develop high-throughput methods, such as PARE (15–18). In this study, a large-scale investigation of in vivo miRNA–target pairs was performed based on the public PARE datasets. For this purpose, the PARE short reads were mapped to the mRNAs. The signatures containing overlapping regions with the predicted target sites were considered to be potential cleavage signals that support the specific miRNA–target regulatory relationships. Besides, the sample origin and the normalized expression level (in RPM) of each short read are provided for reference. Thus, the functional module, ‘MiR–Tar’, was established (Figure 2B).
Theoretically, 3′ remnants generated after the pri-miRNA processing by Dicer-like 1 (DCL1) can be included in plant degradome libraries and detected by PARE sequencing (17,24). Thus, the DCL1-mediated first-step cleavages on both strands of the stem region of a specific pri-miRNA, which result in two poly(A)-tailed remnants, can be validated by PARE data. On the other hand, mature miRNA, and occasionally its miRNA*, can recognize their host precursor as a target based on the complementary sequence present in the stem region. This type of miRNA- or miRNA*-mediated self-regulation can be also represented by plant degradome PARE sequencing data (15,24). Therefore, the module, ‘Self-reg’, was created in PmiRKB to provide a comprehensive view of miRNA precursor processing and miRNA- or miRNA*-mediated self-cleavages on their host precursors (Figure 2C). Similar to the ‘MiR–Tar’ module, the sample origin and the normalized expression level (in RPM) of each short read are provided for reference.
PmiRKB additionally includes the module, ‘MiR info’, which provides sequences of mature miRNAs and pre-miRNAs in FASTA format, clustered pre-miRNAs (within 50 kb), experimental evidences, references and external links to miRBase (6) and PMRD (7). Several useful web links for accessing miRNA-related data and additional analytical tools are provided by ‘Useful links’. Beginners may also appreciate the resources provided in ‘Instructions’ and ‘References’ (Figure 1).
DATABASE IMPLEMENTATION
PmiRKB, with web pages written in PHP scripting language, is maintained on an Apache2 HTTP server containing a PHP5 module. The connection between PmiRKB and the PostgreSQL 8.4 database, containing all of the analytical results, was accomplished by the PostgreSQL client extension of PHP5. Dynamic graphics provided in the four major modules were implemented by SVG, while a few Javascript codes were employed for the dynamic effects experienced on the web pages, such as the movable windows.
FEATURES
Although huge quantities of plant SNP data are currently available, few analyses have been applied for a large-scale investigation of their biological significances for specific types of genes, such as miRNAs. Based on the analytical results accessible from our PmiRKB, SNPs with the potential to alter the secondary structures of pre-miRNAs can be identified. Furthermore, the SNPs within the predicted miRNA–target duplexes, which may interrupt miRNA–target regulation, were also included in the ‘SNP’ module of PmiRKB.
Technological advances in sequencing have greatly accelerated the studies on plant transcriptomes (5). However, analyses of these newly generated HTS data remain incomplete. Here, the ingenious use of HTS data derived from poly(A)-tailed transcripts enabled us to uncover the transcriptional contexts of pri-miRNAs. Considering the lack of information regarding pri-miRNAs in the current miRNA databases, the ‘Pri-miR’ module is a valuable resource for the study of plant miRNA transcription. Specifically, it can facilitate primer or probe designing for pri-miRNA detection. Besides, the level of a specific pri-miRNA will not only indicate the extent of miRNA transcription, but also the stability of the pri-miRNA in vivo. Thus, ‘Pri-miR’ will also provide valuable insights into the efficiency of pri-miRNA processing in a specific tissue.
Another valuable resource provided by PmiRKB is the whole-transcriptome identification of miRNA–target pairs in planta, based on the PARE data (15–17). For researchers who intend to validate miRNA–target pairs of interest, the ‘MiR–Tar’ module can provide the information of cleavage signal intensities. Thus, the priority of each miRNA–target pair to be validated could be determined before modified 5′ RACE assays. A sizable portion of 3′ cleavage remnants, especially those at trace levels, are difficult to be detected by the traditional RACE method. Instead, they could be reflected by PARE data, considering the in-depth feature of HTS technology. Together, the ‘MiR–Tar’ module provides a comprehensive set of miRNA–target pair candidates, including novel pairs with subtle regulatory relationships. Finally, the ‘Self-reg’ module provides novel insights into miRNA precursor processing and miRNA- or miRNA*-mediated self-regulation of miRNA genes.
FUTURE DEVELOPMENT
As the interest in the miRNA world grows, more and more datasets are being generated. Correspondingly, there is an urgent need to organize, parse, and analyze the data produced. This can be partially accomplished through the development of both comprehensive and specialized databases. Here, the creation of a plant miRNA knowledge base, PmiRKB, has been described. The potential for its four functional modules to provide valuable insights into the transcription, processing and regulation of miRNA genes has been demonstrated.
PmiRKB has been updated based on the current release of miRBase (from release 14 to 15) (6). In the near future, more plant species will be included, and PmiRKB will be timely updated following the new release of miRBase (6). In addition, as more HTS data will become publicly available, the results provided by the four modules of PmiRKB could be further refined.
FUNDING
Funding for open access charge: National High Technology Research and Development Program of China (“863” Program) (2008AA10Z125); National Natural Sciences Foundation of China (30771326, 30971743, 31050110121); Program for New Century Excellent Talents in University of China (NCET-07-0740).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Dr Ramanjulu Sunkar for his kindness to provide a free access to the valuable rice degradome sequencing data during the construction of PmiRKB. The authors thank Dr Christian Klukas for his kind discussions. The authors also thank Dr Michael Galperin and the two anonymous referees for their constructive and helpful suggestions.
REFERENCES
- 1.Kim VN, Han J, Siomi MC. Biogenesis of small RNAs in animals. Nat. Rev. Mol. Cell. Biol. 2009;10:126–139. doi: 10.1038/nrm2632. [DOI] [PubMed] [Google Scholar]
- 2.Voinnet O. Origin, biogenesis, and activity of plant microRNAs. Cell. 2009;136:669–687. doi: 10.1016/j.cell.2009.01.046. [DOI] [PubMed] [Google Scholar]
- 3.Jones-Rhoades MW, Bartel DP, Bartel B. MicroRNAs and their regulatory roles in plants. Annu. Rev. Plant Biol. 2006;57:19–53. doi: 10.1146/annurev.arplant.57.032905.105218. [DOI] [PubMed] [Google Scholar]
- 4.Morozova O, Marra MA. Applications of next-generation sequencing technologies in functional genomics. Genomics. 2008;92:255–264. doi: 10.1016/j.ygeno.2008.07.001. [DOI] [PubMed] [Google Scholar]
- 5.Simon SA, Zhai J, Nandety RS, McCormick KP, Zeng J, Mejia D, Meyers BC. Short-read sequencing technologies for transcriptional analyses. Annu. Rev. Plant Biol. 2009;60:305–333. doi: 10.1146/annurev.arplant.043008.092032. [DOI] [PubMed] [Google Scholar]
- 6.Griffiths-Jones S, Saini HK, van Dongen S, Enright AJ. miRBase: tools for microRNA genomics. Nucleic Acids Res. 2008;36:D154–D158. doi: 10.1093/nar/gkm952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang Z, Yu J, Li D, Zhang Z, Liu F, Zhou X, Wang T, Ling Y, Su Z. PMRD: plant microRNA database. Nucleic Acids Res. 2010;38:D806–D813. doi: 10.1093/nar/gkp818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huala E, Dickerman AW, Garcia-Hernandez M, Weems D, Reiser L, LaFond F, Hanley D, Kiphart D, Zhuang M, Huang W, et al. The Arabidopsis Information Resource (TAIR): a comprehensive database and web-based information retrieval, analysis, and visualization system for a model plant. Nucleic Acids Res. 2001;29:102–105. doi: 10.1093/nar/29.1.102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yuan Q, Ouyang S, Liu J, Suh B, Cheung F, Sultana R, Lee D, Quackenbush J, Buell CR. The TIGR rice genome annotation resource: annotating the rice genome and creating resources for plant biologists. Nucleic Acids Res. 2003;31:229–233. doi: 10.1093/nar/gkg059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Clark RM, Schweikert G, Toomajian C, Ossowski S, Zeller G, Shinn P, Warthmann N, Hu TT, Fu G, Hinds DA, et al. Common sequence polymorphisms shaping genetic diversity in Arabidopsis thaliana. Science. 2007;317:338–342. doi: 10.1126/science.1138632. [DOI] [PubMed] [Google Scholar]
- 11.Feltus FA, Wan J, Schulze SR, Estill JC, Jiang N, Paterson AH. An SNP resource for rice genetics and breeding based on subspecies indica and japonica genome alignments. Genome Res. 2004;14:1812–1819. doi: 10.1101/gr.2479404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.McNally KL, Childs KL, Bohnert R, Davidson RM, Zhao K, Ulat VJ, Zeller G, Clark RM, Hoen DR, Bureau TE, et al. Genomewide SNP variation reveals relationships among landraces and modern varieties of rice. Proc. Natl Acad. Sci. USA. 2009;106:12273–12278. doi: 10.1073/pnas.0900992106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.McNally KL, Bruskiewich R, Mackill D, Buell CR, Leach JE, Leung H. Sequencing multiple and diverse rice varieties. Connecting whole-genome variation with phenotypes. Plant Physiol. 2006;141:26–31. doi: 10.1104/pp.106.077313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Nakano M, Nobuta K, Vemaraju K, Tej SS, Skogen JW, Meyers BC. Plant MPSS databases: signature-based transcriptional resources for analyses of mRNA and small RNA. Nucleic Acids Res. 2006;34:D731–D735. doi: 10.1093/nar/gkj077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.German MA, Pillay M, Jeong DH, Hetawal A, Luo S, Janardhanan P, Kannan V, Rymarquis LA, Nobuta K, German R, et al. Global identification of microRNA-target RNA pairs by parallel analysis of RNA ends. Nat. Biotechnol. 2008;26:941–946. doi: 10.1038/nbt1417. [DOI] [PubMed] [Google Scholar]
- 16.Zhou M, Gu L, Li P, Song X, Wei L, Chen Z, Cao X. Degradome sequencing reveals endogenous small RNA targets in rice (Oryza sativa L. ssp. indica) Front. Biol. 2010;5:67–90. [Google Scholar]
- 17.Li YF, Zheng Y, Addo-Quaye C, Zhang L, Saini A, Jagadeeswaran G, Axtell MJ, Zhang W, Sunkar R. Transcriptome-wide identification of microRNA targets in rice. Plant J. 2010;62:742–759. doi: 10.1111/j.1365-313X.2010.04187.x. [DOI] [PubMed] [Google Scholar]
- 18.Addo-Quaye C, Eshoo TW, Bartel DP, Axtell MJ. Endogenous siRNA and miRNA targets identified by sequencing of the Arabidopsis degradome. Curr. Biol. 2008;18:758–762. doi: 10.1016/j.cub.2008.04.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang G, Guo G, Hu X, Zhang Y, Li Q, Li R, Zhuang R, Lu Z, He Z, Fang X, et al. Deep RNA sequencing at single base-pair resolution reveals high complexity of the rice transcriptome. Genome Res. 2010;20:646–654. doi: 10.1101/gr.100677.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhang Y. miRU: an automated plant miRNA target prediction server. Nucleic Acids Res. 2005;33:W701–W704. doi: 10.1093/nar/gki383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hofacker IL. Vienna RNA secondary structure server. Nucleic Acids Res. 2003;31:3429–3431. doi: 10.1093/nar/gkg599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brenner S, Johnson M, Bridgham J, Golda G, Lloyd DH, Johnson D, Luo S, McCurdy S, Foy M, Ewan M, et al. Gene expression analysis by massively parallel signature sequencing (MPSS) on microbead arrays. Nat. Biotechnol. 2000;18:630–634. doi: 10.1038/76469. [DOI] [PubMed] [Google Scholar]
- 23.Lee Y, Kim M, Han J, Yeom KH, Lee S, Baek SH, Kim VN. MicroRNA genes are transcribed by RNA polymerase II. EMBO J. 2004;23:4051–4060. doi: 10.1038/sj.emboj.7600385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meng Y, Gou L, Chen D, Wu P, Chen M. High-throughput degradome sequencing can be used to gain insights into microRNA precursor metabolism. J. Exp. Bot. 2010 doi: 10.1093/jxb/erq209. [Epub ahead of print, July 19, 2010; doi: 10.1093/jxb/erq209] [DOI] [PubMed] [Google Scholar]