Abstract
Mitochondria constitute complex and flexible cellular entities, which play crucial roles in normal and pathological cell conditions. The database MitoGenesisDB focuses on the dynamic of mitochondrial protein formation through global mRNA analyses. Three main parameters confer a global view of mitochondrial biogenesis: (i) time-course of mRNA production in highly synchronized yeast cell cultures, (ii) microarray analyses of mRNA localization that define translation sites and (iii) mRNA transcription rate and stability which characterize genes that are more dependent on post-transcriptional regulation processes. MitoGenesisDB integrates and establishes cross-comparisons between these data. Several model organisms can be analyzed via orthologous relationships between interspecies genes. More generally this database supports the ‘post-transcriptional operon’ model, which postulates that eukaryotes co-regulate related mRNAs based on their functional organization in ribonucleoprotein complexes. MitoGenesisDB allows identifying such groups of post-trancriptionally regulated genes and is thus a useful tool to analyze the complex relationships between transcriptional and post-transcriptional regulation processes. The case of respiratory chain assembly factors illustrates this point. The MitoGenesisDB interface is available at http://www.dsimb.inserm.fr/dsimb_tools/mitgene/.
INTRODUCTION
Mitochondrial biogenesis is an elaborate cellular process that relies on the tight linking of various regulatory controls, from nuclear transcription of genes to the site specific-production of proteins (1,2). Fundamental questions about the determination of the spatio-temporal rules governing the association of the mitochondrial proteins into functional complexes have been largely addressed in the literature. Most of the studies use genetic and biochemical approaches to focus on a few mitochondrial complexes [for instance (3,4)]. In sharp contrast with these analyses, other works provide genome-wide data that give a more comprehensive view of the gene expression program governing mitochondrial biogenesis (1,5–7). In yeast Saccharomyces cerevisiae (S. cerevisiae), the coordinated association of more than 800 proteins (mostly encoded by the nuclear genome) are required to assemble a functional organelle (8,9). To better understand the biology underlying such a complex process, aggregation of multiple sources of genome-wide information is an interesting approach. In this context, data mining constitutes a well-recognized challenge, especially when the data are scattered among different publications and websites.
We present here MitoGenesisDB, a database that offers an easy method to mine and visualize information obtained with global mRNA analyses in the yeast S. cerevisiae. MitoGenesisDB couples data mining tools with a user-friendly web interface so that, with a few mouse clicks, on can easily obtain a rough snapshot of the transcriptome state during mitochondrial biogenesis, in term of (i) mRNA production (5,6), (ii) mRNA cellular localization (1) and (iii) mRNA stability (7). The database can be searched either by specifying a particular gene list, by selecting a specific mitochondrial function or by entering one or several keywords. Orthologous relationships between S. cerevisiaie and other model organisms (Human, Mus Musculus, Arabidopsis thaliana and Caenorhabditis elegans) are supplied in order to enable the database exploration for multiple species. Graphical representations are provided to visualize the results in the context of current biological knowledge and finally, summary page for each gene is proposed with external links to reference databases such as the Saccharomyces Genome Database (SGD) (10) and the Ensembl database (11). The philosophy of MitoGenesisDB is to empower biologists by providing a straightforward data mining interface, and by generating easily interpretable graphical outputs. This should help to mine genome-wide data and supply new openings for the global study of mitochondrial biogenesis.
DATA SETS AVAILABLE IN MITOGENESISDB
Mitochondrial functions
The MitoGenesisDB database contains general information for all the genomic features recorded in the SGD (10) (6.667 features in June 2010). Data stored are the systematic name, the standard name and a general description. From all these features, 794 are genes identified by Saint-Georges et al. (1) as being involved in mitochondrial biogenesis. In MitoGenesisDB, they were manually clustered into eleven model functional groups, related to mitochondria. These groups are labeled ‘Amino Acid Synthesis’; ‘Assembly Factors’; ‘Fe-S Clusters’; ‘Metabolism’; ‘Morphology’; ‘Protein Import’; ‘Respiratory Chain Complexes’; ‘TCA Cycle’; ‘Transport’; ‘Translation Machinery’ and ‘Translation Regulation’ (see the documentation available online for a detailed list of genes attributed to each functional group).
Time-course of mRNA production in highly synchronized yeast cell cultures
In order to confer a global view of mitochondrial biogenesis, we collected microarray data from the study of Tu et al. (5) [accession number GSE3431 in the Gene Expression Omnibus (GEO) database (12)]. The authors used a yeast system with synchronous properties and observed physiological metabolic cycles in connection with a periodicity in the genome expression. Notably most of the genes associated with mitochondria appeared to be expressed with exceptionally robust periodicity. Recently, we developed an original algorithm (called EDPM for Expression Decomposition Based on Periodic Models) to analyze in more details these oscillatory patterns (6). We were able to distinguish six clusters labeled A to F. They comprise distinct subclass of mitochondrial genes for which mRNAs peak in different time window of the metabolic cycles. The temporal groups A to F correlate with functional properties of the corresponding proteins. The first mRNAs to appear are those for genes whose function is associated with translation machinery (or regulation) and assembly factors, followed by those involved in the synthesis of respiratory chain structural proteins and finally mRNAs coding for enzymes involved in the amino-acid biosynthesis. Microarray data for all the genomic features analyzed in Tu et al. (5) (6.551 features) and EDPM results obtained for all the genes analyzed in Lelandais et al. (6) (626 genes) are stored in MitoGenesisDB.
Global analyses of mRNA localization that define translation sites
Other interesting data were collected from the publication of Saint-Georges et al. (1). In this study, the authors quantified for all the genes involved in mitochondrial biogenesis, the Mitochondrial Localization of mRNA (MLR) using microarray experiments and statistical FISH analyses. Three classes of nuclear mRNAs were reported. Classes I and II mRNAs are found near mitochondria, whereas Class III mRNAs are translated on free cytoplamic polysomes. Distinction between Classes I and II mRNAs deals with their subcellular localization: Class I mRNAs is dependent on the activity of the RNA binding protein Puf3p, whereas Class II mRNAs is Puf3p independent. Notably coordination between mRNA oscillations (see previous section) and translation sites in the cell was observed (6). Class I mRNAs dominate in the EDPM cluster A, whereas Classes II and III mRNAs are more evenly distributed among the other clusters. MLR values and MLR classes for all the genes analyzed in Saint-Georges et al. (1) (794 genes) are stored in MitoGenesisDB.
Global mRNA analyses to evaluate the balance between transcriptional and post-transcriptional controls
Previous data sets demonstrate that mitochondrial biogenesis involves a precise coordination between the time at which mRNAs are produced and their final localization in the cell. This coordination needs, on the one hand, transcriptional control, and on the other hand, post-transcriptional regulatory processes. To estimate the balance between these two cellular controls, we collected genome-wide data related to transcription rate and mRNA stability. In Garcia-Martinez et al. (7), the authors used macroarray experiments to calculate for each gene a ‘r coefficient’ that estimates the correlation between values of transcription rate and mRNA levels. To summarize, the r coefficient reflects the global nature of gene regulation. A positive value highlights the role of the transcription rate, whereas a negative value underscores the importance of post-transcriptional processes. Especially, many mitochondrial proteins have negative r coefficients suggesting an important role for post-transcriptional regulatory controls. Such a result agrees with our previous observations that transcriptional and post-transcriptional regulations alternate through the mitochondrial cycle (6). R coefficients for all the genes analyzed in Garcia-Martinez et al. (7) (5.276 genes) are stored in MitoGenesisDB.
GENERAL USE OF MITOGENESISDB
Availability and technical information
MitoGenesisDB is available at http://www.dsimb.inserm.fr/dsimb_tools/mitgene/. It is composed of three parts: a relational database storing information collected from different publications (see the previous section), a web-interface and a set of programs to dynamically generate result files and graphical representations. All the softwares used to power MitoGenesisDB are freely distributed under an open source licence. Data sets have been stored in a MySQL database, the interface has been written in PHP and PERL, with HTML and CSS for page presentation. Graphical outputs are dynamically generated using R programming language.
Main features of MitoGenesisDB
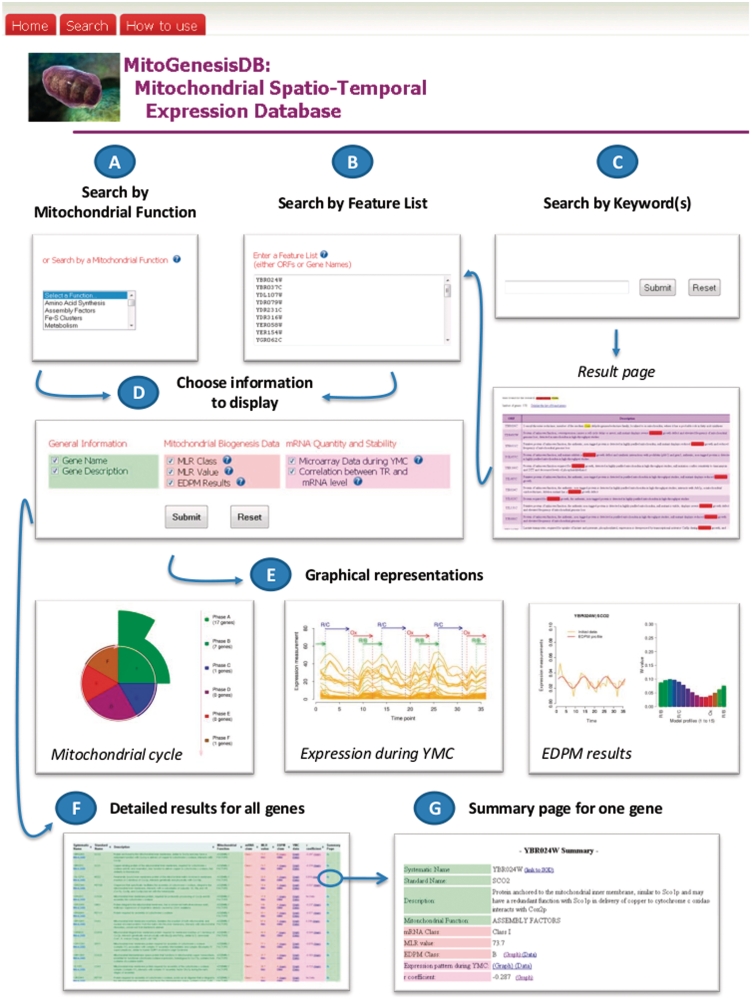
The main features of the MitoGenesisDB are presented Figure 1. The interrogation forms (Figure 1A–C) allow the selection of a list of genes to be queried. A filter option enables to select the data sets to be investigated (Figure 1D), thus allowing to restrict data exploration according to one’s criteria. Comprehensive graphical representations are provided (Figure 1E) to visualize and summarize the results obtained for the requested list of genes. For instance, we provide a graphical representation of the mitochondrial cycle, i.e. a pie chart that shows the correspondence between the different EDPM clusters (6) and the major R/B, R/C and Ox phases identified in the 5-h (or 300-min) yeast metabolic cycle (YMC) (5). Results obtained for each gene are also reported in a table (Figure 1F), where links to summary pages are provided (Figure 1G). Note that the result table can be downloaded in a text format for further examinations with other tools.
Figure 1.
Main features of MitoGenesisDB. The upper part of this figure indicates three major ways to use MitoGenesisDB. The database can be searched (A) by specifying a particular mitochondrial function, (B) by specifying a particular gene list and (C) by entering one or several keywords. (D) Different types of information related to mRNA global analyses can be displayed. (E) Graphical representations are provided to visualize the results of the database queries in the context of current biological knowledge. (F and G) Additional information is also available with for instance, external links to the individual gene description pages of the SGD (10).
Multi-species exploration via orthologous gene lists
All the information stored in the database MitoGenesisDB was obtained in the model yeast S. cerevisiae. To allow the analysis of genes from other model species (Human, Mus Musculus, Arabidopsis Thaliana and Caenorhabditis elegans) we implemented a specific module for orthologue conversion. The main idea is to convert gene names of other species into their orthologous counterpart in S. cerevisiae. For that, we use orthologous relationships available in the INPARANOID database (13). Once the conversion into S. cerevisiae genes is performed, the list can be directly posted into the MitoGenesisDB access ‘Search by Feature List’ (Figure 1B).
A TYPICAL ANALYSIS: THE CASE OF THE RESPIRATORY CHAIN ASSEMBLY FACTORS FAMILY
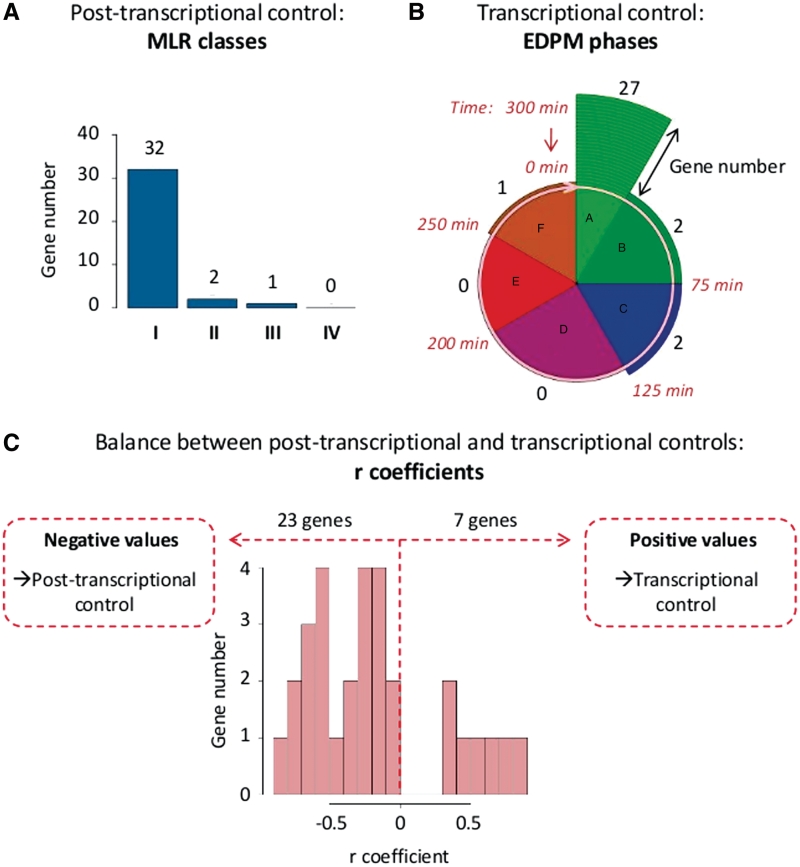
Oxidative phosphorylation is the metabolic pathway used to synthesize adenosine triphosphate (ATP). This process occurs in mitochondria and involves a complex machinery composed of five multi-subunit inner membrane-embedded complexes (the respiratory chain and the ATP synthase), and is built up of more than 90 protein subunits. In the budding yeast S. cerevisiae, the correct assembly of the entire system required time-controlled processes that rely on, at least, 35 assembly factors (see the documentation available online for a detailed description of these 35 genes). As they stimulate and control specific steps of protein complex assembly, the assembly factor production has to be tightly regulated. Curiously enough the genes coding for these factors are not transcriptionally regulated (14,15). When these 35 genes were examined with MitoGenesisDB, several common features revealed interesting new properties, relevant with their regulation process (Figure 2). First, 32/35 of their mRNAs are Class I (Figure 2A). This observation implies that they are translated to the vicinity of mitochondria and that this localization is dependent on the mRNA binding protein Puf3p. Second, a large majority of their mRNAs (27/35) are more present during EDPM phase A, that is a short period (25 min) at the early stage of the metabolic cycles (Figure 2B). Third, 23/30 have negative r coefficient (there are missing values for five genes). The negative correlation between transcription rate and mRNA level of these transcripts reflects a predominant post-transcriptional regulation process (Figure 2C).
Figure 2.
Example of study. MitoGenesisDB was used to analyze the respiratory chain assembly factors family (35 genes). (A) Distribution of genes in MLR classes as defined in Saint-Georges et al. (1). 32/35 genes belong to the MLR Class I, meaning that these genes have transcripts located at the vicinity of mitochondria and this localization is dependent on the Puf3p protein. (B) mRNA quantity of genes during the YMC. This pie chart shows the correspondence between the different EDPM classes (phase A to F) identified in Lelandais et al. (6) and the time points during the YMC (from 0 to 300 min) identified in Tu et al. (5). The number of genes in each EDPM class is represented with surrounding circular segments. 27/35 of the transcripts are present in phase A that is the early stage of the YMC. (C) Histogram of the r coefficients as defined in Garcia-Martinez et al. (7). 23/30 have negative r coefficients (no value was available for five genes). Such an observation underscores the importance of post-transcriptional processes.
All together, these observations suggest that assembly factors belong to a same group of spatio-temporal expression. This rather clear-cut observation that was not expected and it raises several interesting questions. For instance, how do assembly factors control the early steps of respiratory chain biogenesis and how can we explain the predominant role of a synchronized post-transcriptional control in their regulation? Do they control the topologic sites where the respiratory complexes are constructed? Are they connected to the biogenesis of mitochondrial-encoded subunits which constitute the core complexes? Further experiments are needed to answer these challenging questions, but the use of a database like MitoGenesisDB represent a good starting point.
CONCLUSION AND FUTURE DEVELOPMENTS
With MitoGenesisDB, our aim is to take advantage of genome-wide data sets to better understand the spatio-temporal regulation of mitochondrial biogenesis. Several regulatory levels, from transcriptional to post-transcriptional processes, can be explored through the association of information related to mRNAs production, mRNAs cellular localization and r coefficients to evaluate the balance between transcriptional and post-transcriptional regulatory controls. The user-friendly web interface is designed to be accessible to those with no particular technical skill, and graphical outputs are provided allowing the user to elaborate rapidly his (or her) own interpretation of the data.
Compared to the existing tools in the field like MitoP2 (16), MitoDat (17), MitoRes (18), Mitodrome (19), MitoMiner (20), Mitomap (21) or Mitome (22), MitoGenesisDB is the first database that integrates results obtained with global transcriptome analyses. The major drawback of classical mRNAs analyses is that coordinated waves of transcription/translation are difficult to observe because of the metabolic asynchrony of the cells in growing cultures. In MitoGenesisDB, we provide expression data obtained from yeasts grown under continuous and nutrient-limited conditions, and in which cell-to-cell signaling synchronizes metabolic functions (5). The gene-expression dynamic of the YMC is therefore a useful model system to gain a comprehensive picture of the biogenesis of yeast mitochondria. More generally, as it underlines temporal differences between clusters of co-expressed genes (6), we believe that the YMC is an interesting model for studies of the lifecycle of any groups of transcripts in eukaryotic cells (23).
At present, the interpretation of the MitoGenesisDB results obtained for other species than yeast is limited, because of the gene conversion via orthologous links with S. cerevisiae. A natural future direction for the database development is to incorporate experimental data directly originated from multiple organisms. Also the addition of information related to 3′ and 5′ regulatory elements in mRNA UTR sequences is a promising perspective to better investigate the regulatory processes governing the tight coordination between transcriptional and post-transcriptional processes involved in mitochondrial biogenesis.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Institut National de la Transfusion Sanguine (INTS).
Conflict of interest statement. None declared.
REFERENCES
- 1.Saint-Georges Y, Garcia M, Delaveau T, Jourdren L, Le Crom S, Lemoine S, Tanty V, Devaux F, Jacq C. Yeast mitochondrial biogenesis: a role for the PUF RNA-binding protein Puf3p in mRNA localization. PLoS ONE. 2008;3:e2293. doi: 10.1371/journal.pone.0002293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Garcia M, Delaveau T, Goussard S, Jacq C. Mitochondrial presequence and open reading frame mediate asymmetric localization of messenger RNA. EMBO Rep. 2010;11:285–291. doi: 10.1038/embor.2010.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Fontanesi F, Soto IC, Horn D, Barrientos A. Assembly of mitochondrial cytochrome c-oxidase, a complicated and highly regulated cellular process. Am. J. Physiol. Cell Physiol. 2006;291:C1129–C1147. doi: 10.1152/ajpcell.00233.2006. [DOI] [PubMed] [Google Scholar]
- 4.Garcia M, Darzacq X, Delaveau T, Jourdren L, Singer RH, Jacq C. Mitochondria-associated yeast mRNAs and the biogenesis of molecular complexes. Mol. Biol. Cell. 2007;18:362–368. doi: 10.1091/mbc.E06-09-0827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tu BP, Kudlicki A, Rowicka M, McKnight SL. Logic of the yeast metabolic cycle: temporal compartmentalization of cellular processes. Science. 2005;310:1152–1158. doi: 10.1126/science.1120499. [DOI] [PubMed] [Google Scholar]
- 6.Lelandais G, Saint-Georges Y, Geneix C, Al-Shikhley L, Dujardin G, Jacq C. Spatio-temporal dynamics of yeast mitochondrial biogenesis: transcriptional and post-transcriptional mRNA oscillatory modules. PLoS Comput. Biol. 2009;5:e1000409. doi: 10.1371/journal.pcbi.1000409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Garcia-Martinez J, Aranda A, Perez-Ortin JE. Genomic run-on evaluates transcription rates for all yeast genes and identifies gene regulatory mechanisms. Mol. Cell. 2004;15:303–313. doi: 10.1016/j.molcel.2004.06.004. [DOI] [PubMed] [Google Scholar]
- 8.Perocchi F, Jensen LJ, Gagneur J, Ahting U, von Mering C, Bork P, Prokisch H, Steinmetz LM. Assessing systems properties of yeast mitochondria through an interaction map of the organelle. PLoS Genet. 2006;2:e170. doi: 10.1371/journal.pgen.0020170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Elstner M, Andreoli C, Ahting U, Tetko I, Klopstock T, Meitinger T, Prokisch H. MitoP2: an integrative tool for the analysis of the mitochondrial proteome. Mol. Biotechnol. 2008;40:306–315. doi: 10.1007/s12033-008-9100-5. [DOI] [PubMed] [Google Scholar]
- 10.Christie KR, Weng S, Balakrishnan R, Costanzo MC, Dolinski K, Dwight SS, Engel SR, Feierbach B, Fisk DG, Hirschman JE, et al. Saccharomyces Genome Database (SGD) provides tools to identify and analyze sequences from Saccharomyces cerevisiae and related sequences from other organisms. Nucleic Acids Res. 2004;32:D311–D314. doi: 10.1093/nar/gkh033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Edgar R, Domrachev M, Lash AE. Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res. 2002;30:207–210. doi: 10.1093/nar/30.1.207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fontanesi F, Soto IC, Barrientos A. Cytochrome c oxidase biogenesis: new levels of regulation. IUBMB Life. 2008;60:557–568. doi: 10.1002/iub.86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Barrientos A, Fontanesi F, Diaz F. Evaluation of the mitochondrial respiratory chain and oxidative phosphorylation system using polarography and spectrophotometric enzyme assays. Curr. Protoc. Hum. Genet. 2009 doi: 10.1002/0471142905.hg1903s63. Chapter 19, Unit19 13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Prokisch H, Andreoli C, Ahting U, Heiss K, Ruepp A, Scharfe C, Meitinger T. MitoP2: the mitochondrial proteome database–now including mouse data. Nucleic Acids Res. 2006;34:D705–D711. doi: 10.1093/nar/gkj127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Lemkin PF, Chipperfield M, Merril C, Zullo S. A World Wide Web (WWW) server database engine for an organelle database, MitoDat. Electrophoresis. 1996;17:566–572. doi: 10.1002/elps.1150170327. [DOI] [PubMed] [Google Scholar]
- 18.Catalano D, Licciulli F, Turi A, Grillo G, Saccone C, D'Elia D. MitoRes: a resource of nuclear-encoded mitochondrial genes and their products in Metazoa. BMC Bioinformatics. 2006;7:36. doi: 10.1186/1471-2105-7-36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sardiello M, Licciulli F, Catalano D, Attimonelli M, Caggese C. MitoDrome: a database of Drosophila melanogaster nuclear genes encoding proteins targeted to the mitochondrion. Nucleic Acids Res. 2003;31:322–324. doi: 10.1093/nar/gkg123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Smith AC, Robinson AJ. MitoMiner, an integrated database for the storage and analysis of mitochondrial proteomics data. Mol. Cell Proteomics. 2009;8:1324–1337. doi: 10.1074/mcp.M800373-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ruiz-Pesini E, Lott MT, Procaccio V, Poole JC, Brandon MC, Mishmar D, Yi C, Kreuziger J, Baldi P, Wallace DC. An enhanced MITOMAP with a global mtDNA mutational phylogeny. Nucleic Acids Res. 2007;35:D823–D828. doi: 10.1093/nar/gkl927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee YS, Oh J, Kim YU, Kim N, Yang S, Hwang UW. Mitome: dynamic and interactive database for comparative mitochondrial genomics in metazoan animals. Nucleic Acids Res. 2008;36:D938–D942. doi: 10.1093/nar/gkm763. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Palumbo MC, Farina L, De Santis A, Giuliani A, Colosimo A, Morelli G, Ruberti I. Collective behavior in gene regulation: post-transcriptional regulation and the temporal compartmentalization of cellular cycles. FEBS J. 2008;275:2364–2371. doi: 10.1111/j.1742-4658.2008.06398.x. [DOI] [PubMed] [Google Scholar]