Abstract
Transient receptor potential (TRP) channels are a superfamily of Ca2+-permeable cation channels that translate cellular stimuli into electrochemical signals. Aberrant activity of TRP channels has been implicated in a variety of human diseases, such as neurological disorders, cardiovascular disease and cancer. To facilitate the understanding of the molecular network by which TRP channels are associated with biological and disease processes, we have developed the TRIP (TRansient receptor potential channel-Interacting Protein) Database (http://www.trpchannel.org), a manually curated database that aims to offer comprehensive information on protein–protein interactions (PPIs) of mammalian TRP channels. The TRIP Database was created by systematically curating 277 peer-reviewed literature; the current version documents 490 PPI pairs, 28 TRP channels and 297 cellular proteins. The TRIP Database provides a detailed summary of PPI data that fit into four categories: screening, validation, characterization and functional consequence. Users can find in-depth information specified in the literature on relevant analytical methods and experimental resources, such as gene constructs and cell/tissue types. The TRIP Database has user-friendly web interfaces with helpful features, including a search engine, an interaction map and a function for cross-referencing useful external databases. Our TRIP Database will provide a valuable tool to assist in understanding the molecular regulatory network of TRP channels.
INTRODUCTION
Transient receptor potential (TRP) channels are a large family of Ca2+-permeable cation channels that encompass 28 isotypes in mammals (1–5). The TRP channel superfamily is divided into six subfamilies (1–5): TRPC (canonical), TRPV (vanilloid), TRPM (melastatin), TRPP (polycystin), TRPML (mucolipin) and TRPA (ankyrin). TRP channels convert a wide range of environmental stimuli into electrochemical signals via membrane potential and intracellular Ca2+. These common transductions play crucial roles in many physiological processes (6–8). A detailed explanation of the role of TRP channels in physiology is beyond the scope of this paper, and the reader is referred to the excellent review articles found in refs (1–8) and ‘Further Reading’ on the main page of the TRIP Database website.
Accumulating evidence indicates that aberrant TRP channels have causal roles in various human diseases. Mutations in these channels are increasingly linked to genetic disorders, such as focal segmental glomerulosclerosis, mucolipidosis type IV and autosomal dominant polycystic kidney disease (9,10). In addition, deregulation of TRP channel activity is associated with various human diseases, including neurological disorders, cardiovascular disease and cancer (11–13). Furthermore, studies of mice with ablated TRP channels provide insight into the relationship between TRP channel functionality and disease processes (14,15). Thus, TRP channels would make attractive targets for therapeutic intervention. However, the molecular mechanisms by which TRP channels are involved in the diseases are largely unknown.
Most proteins form complexes to achieve specific functions in virtually all biological systems. Correctly identifying and characterizing the protein–protein interactions (PPIs) and the networks they comprise within these complexes are crucial for understanding the molecular mechanisms determining biological phenomena and disease processes (16–20). Recently, PPI interfaces have emerged as promising drug targets (21). In addition, the PPI data gathered comprise frameworks for system-based drug discovery (22–23). In the field of TRP channel research, the publication of PPI papers has markedly increased in recent years (Supplementary Figure S1). Therefore, there is a need to map the molecular landscape of TRP channel PPIs to assist those seeking to gain insight into the molecular mechanisms by which TRP channels are associated with a variety of diseases. However, the ongoing accumulation of information on TRP channel PPIs is scattered.
Enormous information on the global PPI network is hosted by several existing PPI databases, such as DIP (24), IntAct (25), MINT (26), STRING (27) and BioGRID (28). However, these databases do not provide sufficient information to scientists who want to focus their research on particular protein families. Thus, it is desirable to construct databases that contain detailed information on relevant analytical methods, experimental resources and summarized results. Such PPI databases can provide in-depth information on specific molecules or molecule families and will be more effective than the present global PPI network in stimulating the formulation of new knowledge, hypotheses or experiments.
Here, we present the TRIP (TRansient receptor potential channel-Interacting Protein) Database, a manually curated database that aims to provide comprehensive information on PPIs of mammalian TRP channels. The TRIP Database can be accessed at http://www.trpchannel.org.
CONTENTS AND DESIGN
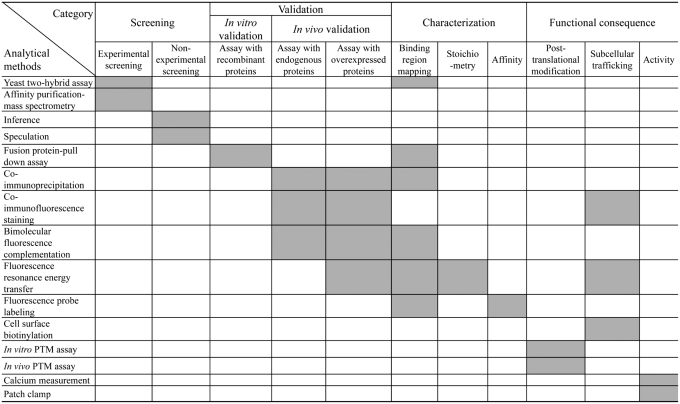
The current version of the TRIP Database documents 490 PPI pairs among 28 TRP channels and 297 cellular proteins (Supplementary Figures S2 and S3). We selected 277 peer-reviewed articles on TRP channel PPIs from a pool of more than 500 to serve as the source of information for the database. An overview of the information described in each article is presented in the form of a matrix (Figure 1). Based on common research schemes of PPI studies (29), columns are labeled with the following categorizations: screening, validation, characterization and functional consequence (Supplementary Table S1). These categorizations give answers to four basic questions about PPIs: how to identify PPIs (screening); how to confirm PPIs (validation); what are the biochemical properties of PPIs (characterization); what is the biological meaning of PPIs (functional consequence). Each category is assigned to the brief summary of experimental results or the information on experimental resources (e.g. gene constructs and cell/tissue types). Rows are labeled with relevant analytical methods as described in the literature (29,30). Based on the information in the ‘Materials and Methods’ section of each paper, we listed 15 analytical methods used to obtain PPI data (Figure 1). In addition, the TRIP Database provides information on the amino acid sequences of TRP channels and their interacting proteins, which is retrieved from the UniProtKB/Swiss-Prot and UniProtKB/TrEMBL databases (31) and available as FASTA, EMBL and GenBank formats (Supplementary Figure S4).
Figure 1.
A detailed summary of TRP channel PPIs provided by the TRIP Database. Analytical methods and their association with categories are represented by gray boxes.
Screening refers to the type of high-throughput experimental methods (i.e. yeast two-hybrid screens or affinity purification followed by mass spectrometry) used to identify the TRP channel-interacting proteins (interactors). Because TRP channels are almost exclusively used as baits in the experiments, we include information on the TRP channel constructs and the target sample sources with regard to species, organ/tissue and sample types as specified in the literature. In addition, we curated the substantial number of TRP channel PPIs discovered by speculation, inference or other non-experimental methods under the category of screening.
Validation is classified either as in vitro or in vivo depending on the experimental techniques used to validate PPIs. In vitro validation refers to the methods that use assays with the recombinant proteins encoded by either TRP channels and/or their interactors (e.g. fusion protein pull-down assays), whereas in vivo validation represents those processes that use assays with endogenous proteins (i.e. assays without transfection) or overexpressed proteins (i.e. assays with transfection). The TRIP Database provides a detailed summary of experimental resources, including the gene constructs and expression systems used for recombinant protein preparation and cell/tissue types.
Characterization refers to the data on the biochemical properties of PPIs, such as binding region mapping, stoichiometry or affinity among interacting proteins as specified in the articles. We curated the mapping data for binding regions with regard to the gene constructs used and amino acid region (e.g. 1–324). In the TRIP Database, stoichiometry is described in quantitative terms (i.e. binding ratio), and affinity is defined by a dissociation constant (i.e. Kd).
Because activity of TRP channels is usually modulated by the changes in post-translational modification (PTM) or subcellular trafficking, we classified the functional consequences described in each article into three subcategories: PTM, subcellular trafficking and activity. We curated the information described in the articles on PTM and subcellular trafficking with regard to the types of PTM (e.g. phosphorylation) and the changes in subcellular trafficking of TRP channels, respectively. In addition, we briefly summarize the experimental results concerning the changes of TRP channel activity.
IMPLEMENTATION
Data collection
The literature on TRP channel PPIs found in the PubMed database serve as the primary information source for constructing the TRIP Database. First, a list of synonyms for the term ‘TRP channels’ was constructed from UniprotKB, Entrez Gene (32), membrane protein databases (Supplementary Table S2) and published review papers for nomenclature (33,34). Second, using these synonyms, a list of articles was obtained through a PubMed search. Third, salient articles were collected through a survey of PubMed abstracts and subsequently by search of full-text papers. Finally, we selected articles that contain evidence for physical binding among the proteins denoted. To prevent omission of relevant papers, we manually screened information in other databases, such as DIP, IntAct, MINT, STRING, BioGRID, Entrez Gene, IUPHAR-DB (35) and ISI Web of Knowledge (from Thomson Reuters). All 277 articles used for database construction are listed in our database website (Supplementary Figure S5). Every entry in our database has corresponding articles for those interested in seeking further information.
Data curation
We curated the data based on analytical methods, experimental results, resources (e.g. genes or proteins, primary cells, tissues and cell lines) and nomenclature. The curation of analytical methods and experimental results was partly described in the earlier section on categorization (see the ‘Contents and design’ section). Regarding nomenclature to describe analytical methods (Figure 1), we employ popularly used names in the papers. However, the names of two methods are arbitrarily designated; one is fusion protein pull down assay, which includes the assays using the recombinant proteins fused with GST, histidine hexamer or other peptide tags; the other is fluorescence probe labeling, which represents the assays using protein labeled with various fluorescence probes.
We manually curated the information on the gene constructs denoted in the literature with respect to species (e.g. mouse) and amino acid region (e.g. 1–324). As with regard to nomenclature to describe the proteins, we adopt common names that are widely used in the literature. If information on the gene constructs was not found in the literature, but relevant references were provided, we gathered information from referenced articles. However, if no appropriate data were available, it is designated as ‘Not specified’ in the TRIP Database.
We also curated nomenclatures for the primary cells, tissues and cell lines used in the validation of TRP channel PPIs. Because the nomenclatures describing primary cells or tissues are mostly unified, we faithfully cited primary cell or tissue names designated in the literature texts. Contrastingly, because the nomenclatures delineating cell lines have not been standardized (36), we sometimes, as previously described, modified the corresponding names by referring to the CLDB (37) and ATCC databases (36).
Database construction and web interface development
The data contained in the TRIP Database are stored on a carefully designed relational database on a MySQL server (version 5.1.41). The web interfaces to the database were developed using XHTML 1.1, CSS, Javascript and Ruby on Rails 2.3.8 technologies. The web application runs on a Phusion Passenger application server (version 2.2.15) with an Apache HTTP server (version 2.2.14) hosted on an Ubuntu Linux server (version 10.0.4 LTS). The web-based visualization tool of the TRIP Database was developed using Adobe Flex 4 SDK and Flare, an open-source data visualization library for Adobe Flash Player.
WEB INTERFACES
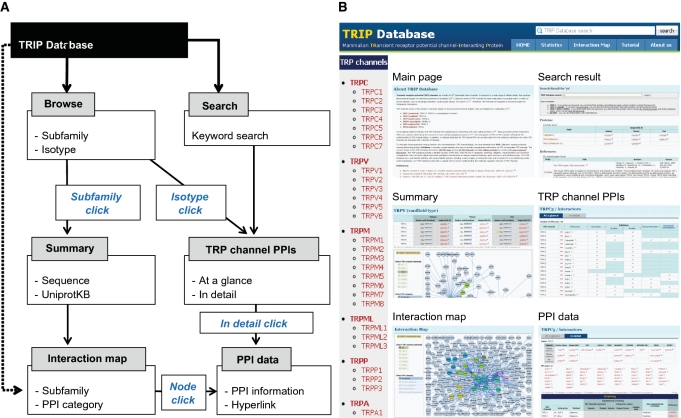
The TRIP Database has user-friendly interfaces with several helpful features, including a search engine, an interaction map and a tool that cross-references other useful external databases. Navigation of the TRIP Database is illustrated in Figure 2, and the website also provides instruction in the ‘Tutorial’ section (see also Supplementary Figure S6). Users can browse the database based on TRP channel subfamilies or isotypes. If users choose to begin their search based on a specific TRP channel subfamily, they will get a summary of the subfamily, including information on isotypes, amino acid sequences or UniprotKB IDs. If users choose to browse based on a specific TRP channel isotype, they will see lists of TRP channel PPI pairs. For example, the interface might provide one TRP channel with all of its interactors whereas in another instance all TRP channels with one interactor.
Figure 2.
Overview of the TRIP Database interfaces. (A) A diagram of navigation of the TRIP Database. (B) Reconstituted illustration based on the navigation.
Search
The TRIP Database provides full-text search capability on proteins, PPIs and relevant literature (Supplementary Figure S7 and Tutorial 5 on our website). When a user enters a query (i.e. protein names or UniprotKB IDs) into the search form, the system presents corresponding information on all proteins and PPI pairs. In addition, the system can provide a list of indexed entries that contain the search word in the title, author, affiliation or abstract.
Interaction map
The TRIP Database provides a visualization tool that graphically displays the network of TRP channel PPIs. Proteins are represented as round-shaped nodes labeled with their names, and PPIs are illustrated as edges connecting the protein nodes (Supplementary Figure S8). By clicking the nodes, users can highlight all PPI pairs generated by a particular protein. Detailed information of a PPI can be obtained by double-clicking the representative node. A control panel on the left side of the toolbar allows users to choose TRP channel subfamilies and PPI categories.
External links
In order to complement the TRIP Database and provide additional information on each gene contained in the TRIP Database, we provide hyperlinks to other useful databases (Supplementary Table S3), including UinprotKB, DIP, IntAct, MINT, STRING, BioGRID, Entrez Gene, IUPHAR-DB, KEGG (38) and OMIM (39). Thus, the TRIP Database works as a central hub for the collection of structural, functional, pharmacological and pathophysiological information on TRP channels and their PPIs.
DISCUSSION AND FUTURE DIRECTIONS
The TRIP Database provides extensive information about TRP channels and their interactions with cellular proteins that allows for in-depth characterization of the molecular network of TRP channels. This database is manually curated and presently contains 490 PPI pair entries. Automated data collection and curation via machine learning techniques often include extraneous data or omit desirable data; therefore, manual curation is currently the best method for constructing reliable biological databases. The superiority of manual curation becomes more obvious when comparing PPI databases (Supplementary Table S4). Compared to our TRIP Database, considerable information on TRP channel PPIs are omitted in other experimentally verified PPI databases. On the other hand, the curator is also able to include all the detailed information, such as species of genes, regions on proteins, or cell/tissue types. Therefore, the TRIP Database will be a useful, convenient and accurate resource for research on TRP channels.
Looking forward, there are several challenges for the TRIP Database. First, a wiki page will be added onto the TRIP Database website to encourage researcher participation. Researchers will be able to comment, add new information or revise the entries in the TRIP Database. We hope to improve the TRIP Database through the collaborating participation of other researchers. Next, we will develop machine learning techniques to cluster new meaningful protein complexes and predict new PPIs. This will stimulate the possible formulation of new hypotheses for the novel role of TRP channels in physiology and provide an integrated view of TRP channel biology. Lastly, we hope to propose a standard format to describe information on TRP channel PPIs. As far as we know, currently there is no standard format to describe information on TRP channel PPIs. The proposed standardized format will aid researchers to exchange or explain their works more effectively and efficiently.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
This research was supported by Basic Science Research Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (2008-05943), and by a grant from the Seoul National University Hospital Research Fund (04-2007-064-0). Y.-C. Shin was supported by graduate program of BK21 project from Ministry of Education, Science and Technology. Funding for open access charge: Ministry of Education, Science and Technology, Graduate program of BK21 project.
Conflict of interest statement. None declared.
REFERENCES
- 1.Ramsey IS, Delling M, Clapham DE. An introduction to TRP channels. Annu. Rev. Physiol. 2006;68:619–647. doi: 10.1146/annurev.physiol.68.040204.100431. [DOI] [PubMed] [Google Scholar]
- 2.Montell C, Birnbaumer L, Flockerzi V. The TRP channels, a remarkably functional family. Cell. 2002;108:595–598. doi: 10.1016/s0092-8674(02)00670-0. [DOI] [PubMed] [Google Scholar]
- 3.Venkatachalam K, Montell C. TRP channels. Annu. Rev. Biochem. 2007;76:387–417. doi: 10.1146/annurev.biochem.75.103004.142819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Owsianik G, Talavera K, Voets T, Nilius B. Permeation and selectivity of TRP channels. Annu. Rev. Physiol. 2006;68:685–717. doi: 10.1146/annurev.physiol.68.040204.101406. [DOI] [PubMed] [Google Scholar]
- 5.Montell C. The TRP superfamily of cation channels. Sci. STKE. 2005;2005:re3. doi: 10.1126/stke.2722005re3. [DOI] [PubMed] [Google Scholar]
- 6.Clapham DE. TRP channels as cellular sensors. Nature. 2003;426:517–524. doi: 10.1038/nature02196. [DOI] [PubMed] [Google Scholar]
- 7.Voets T, Talavera K, Owsianik G, Nilius B. Sensing with TRP channels. Nat. Chem. Biol. 2005;1:85–92. doi: 10.1038/nchembio0705-85. [DOI] [PubMed] [Google Scholar]
- 8.Montell C. Physiology, phylogeny, and functions of the TRP superfamily of cation channels. Sci. STKE. 2001;2001:re1. doi: 10.1126/stke.2001.90.re1. [DOI] [PubMed] [Google Scholar]
- 9.Nilius B, Owsianik G. Transient receptor potential channelopathies. Pflugers Arch. 2010;460:437–450. doi: 10.1007/s00424-010-0788-2. [DOI] [PubMed] [Google Scholar]
- 10.Kiselyov K, Soyombo A, Muallem S. TRPpathies. J. Physiol. 2007;578:641–653. doi: 10.1113/jphysiol.2006.119024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nilius B, Voets T, Peters J. TRP channels in disease. Sci. STKE. 2005;2005:re8. doi: 10.1126/stke.2952005re8. [DOI] [PubMed] [Google Scholar]
- 12.Nilius B. TRP channels in disease. Biochim. Biophys. Acta. 2007;1772:805–812. doi: 10.1016/j.bbadis.2007.02.002. [DOI] [PubMed] [Google Scholar]
- 13.Abramowitz J, Birnbaumer L. Physiology and pathophysiology of canonical transient receptor potential channels. FASEB J. 2009;23:297–328. doi: 10.1096/fj.08-119495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Freichel M, Flockerzi V. Biological functions of TRPs unravelled by spontaneous mutations and transgenic animals. Biochem. Soc. Trans. 2007;35:120–123. doi: 10.1042/BST0350120. [DOI] [PubMed] [Google Scholar]
- 15.Desai BN, Clapham DE. TRP channels and mice deficient in TRP channels. Pflugers Arch. 2005;451:11–18. doi: 10.1007/s00424-005-1429-z. [DOI] [PubMed] [Google Scholar]
- 16.Sharan R, Ulitsky I, Shamir R. Network-based prediction of protein function. Mol. Syst. Biol. 2007;3:88. doi: 10.1038/msb4100129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Legrain P, Wojcik J, Gauthier JM. Protein-protein interaction maps: a lead towards cellular functions. Trends Genet. 2001;17:346–352. doi: 10.1016/s0168-9525(01)02323-x. [DOI] [PubMed] [Google Scholar]
- 18.Drewes G, Bouwmeester T. Global approaches to protein–protein interactions. Curr. Opin. Cell Biol. 2003;15:199–205. doi: 10.1016/s0955-0674(03)00005-x. [DOI] [PubMed] [Google Scholar]
- 19.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein-protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 20.Ideker T, Sharan R. Protein networks in disease. Genome Res. 2008;18:644–652. doi: 10.1101/gr.071852.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wells JA, McClendon CL. Reaching for high-hanging fruit in drug discovery at protein–protein interfaces. Nature. 2007;450:1001–1009. doi: 10.1038/nature06526. [DOI] [PubMed] [Google Scholar]
- 22.Ruffner H, Bauer A, Bouwmeester T. Human protein–protein interaction networks and the value for drug discovery. Drug Discov. Today. 2007;12:709–716. doi: 10.1016/j.drudis.2007.07.011. [DOI] [PubMed] [Google Scholar]
- 23.Fuentes G, Oyarzabal J, Rojas AM. Databases of protein–protein interactions and their use in drug discovery. Curr. Opin. Drug Discov. Dev. 2009;12:358–366. [PubMed] [Google Scholar]
- 24.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The Database of Interacting Proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8-a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID Interaction Database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Xenarios I, Eisenberg D. Protein interaction databases. Curr. Opin. Biotechnol. 2001;12:334–339. doi: 10.1016/s0958-1669(00)00224-x. [DOI] [PubMed] [Google Scholar]
- 30.Shoemaker BA, Panchenko AR. Deciphering protein–protein interactions: Part I. Experimental techniques and databases. PLoS Comput. Biol. 2007;3:e42. doi: 10.1371/journal.pcbi.0030042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Clapham DE, Montell C, Schultz G, Julius D. International Union of Pharmacology: XLIII. Compendium of voltage-gated ion channels: transient receptor potential channels. Pharmacol. Rev. 2003;55:591–596. doi: 10.1124/pr.55.4.6. [DOI] [PubMed] [Google Scholar]
- 34.Clapham DE, Julius D, Montell C, Schultz G. International Union of Pharmacology: XLIX. Nomenclature and structure-function relationships of transient receptor potential channels. Pharmacol. Rev. 2005;57:427–450. doi: 10.1124/pr.57.4.6. [DOI] [PubMed] [Google Scholar]
- 35.Harmar AJ, Hills RA, Rosser EM, Jones M, Buneman OP, Dunbar DR, Greenhill SD, Hale VA, Sharman JL, Bonner TI, et al. IUPHAR-DB: the IUPHAR database of G protein-coupled receptors and ion channels. Nucleic Acids Res. 2009;37:D680–D685. doi: 10.1093/nar/gkn728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sarntivijai S, Ade AS, Athey BD, States DJ. A bioinformatics analysis of the cell line nomenclature. Bioinformatics. 2008;24:2760–2766. doi: 10.1093/bioinformatics/btn502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Romano P, Manniello A, Aresu O, Armento M, Cesaro M, Parodi B. Cell Line Data Base: structure and recent improvements towards molecular authentication of human cell lines. Nucleic Acids Res. 2009;37:D925–D932. doi: 10.1093/nar/gkn730. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]