Abstract
MatrixDB (http://matrixdb.ibcp.fr) is a freely available database focused on interactions established by extracellular proteins and polysaccharides. Only few databases report protein–polysaccharide interactions and, to the best of our knowledge, there is no other database of extracellular interactions. MatrixDB takes into account the multimeric nature of several extracellular protein families for the curation of interactions, and reports interactions with individual polypeptide chains or with multimers, considered as permanent complexes, when appropriate. MatrixDB is a member of the International Molecular Exchange consortium (IMEx) and has adopted the PSI-MI standards for the curation and the exchange of interaction data. MatrixDB stores experimental data from our laboratory, data from literature curation, data imported from IMEx databases, and data from the Human Protein Reference Database. MatrixDB is focused on mammalian interactions, but aims to integrate interaction datasets of model organisms when available. MatrixDB provides direct links to databases recapitulating mutations in genes encoding extracellular proteins, to UniGene and to the Human Protein Atlas that shows expression and localization of proteins in a large variety of normal human tissues and cells. MatrixDB allows researchers to perform customized queries and to build tissue- and disease-specific interaction networks that can be visualized and analyzed with Cytoscape or Medusa.
INTRODUCTION
The extracellular matrix is comprised of proteins and complex polysaccharides that are organized in a tissue-specific manner. Major components of the extracellular matrix are collagens [∼30% of proteins in humans; (1)], elastic fibers, proteoglycans and glycosaminoglycans. Several extracellular protein families (e.g. collagens, laminins and thrombospondins) form stable multimers in their native state, the multimers being comprised of either identical or different polypeptide chains. The extracellular matrix provides a structural scaffold contributing to the mechanical properties of tissues (2), and is a reservoir of bioactive fragments, called matricryptins, that are released upon limited proteolysis. These fragments exhibit biological and biomolecular recognition properties of their own and regulate a number of physiological and pathological processes including angiogenesis and tumor growth (3). The cohesion of the extracellular matrix is maintained by an intricate interaction network of protein–protein and protein–glycosaminoglycan interactions. These interactions are involved in the formation of supramolecular assemblies such as collagen fibrils and elastic fibers, in tissue architecture, and in cell-matrix interactions that regulate cell growth and behavior. The perturbation of the extracellular interaction network by mutations in genes coding for extracellular proteins lead to several diseases ranging from mild to severe phenotypes [e.g. osteogenesis imperfecta; (4)].
Interactions involving extracellular proteins are poorly represented in existing databases, and protein–glycosaminoglycan interactions are almost absent from databases although they contribute to the structural organization of the extracellular matrix, to the sequestration of growth factors and chemokines within the extracellular matrix, and to signalling at the cell surface (5). Furthermore, interactions involving multimers, which are frequent in the extracellular matrix (collagens, laminins, thrombospondins are trimers), are often reported as interactions established by individual polypeptide chains. This is a concern especially when molecules are heteromultimers. The above reasons prompted us to build an interaction database focused on interactions occurring between extracellular biomolecules [http://matrixdb.ibcp.fr; (6)]. The database has been updated to include additional interaction data, comprehensive extracellular interaction datasets (e.g. the elastic fiber interactome, extracellular interactions of leucine-rich repeat receptors), and new functionalities. MatrixDB is focused on mammalian molecules, but interaction data of a model organism (zebrafish) has been integrated in the updated database. MatrixDB provides direct links to Online Mendelian Inheritance in Man (OMIM), to databases recapitulating data on mutations occurring in genes encoding extracellular proteins, to UniGene and to the Human Protein Atlas that shows expression and localization of proteins in a large variety of normal human tissues, cancer cells and cell lines. MatrixDB allows researchers to perform customized queries and to build tissue- and disease-specific interaction networks.
BIOMOLECULE DATA
We have imported protein data from the UniProtKB/Swiss-Prot knowledgebase (7), and used UniProtKB accession numbers for proteins. We have created specific identifiers for multimers such as collagens, laminins, thrombospondins and integrins using the following format: MULT_x_species (e.g. MULT_3_human for human collagen I). These entries refer to the UniProtKB accession numbers of their constituent polypeptide chains. Complexes corresponding to stable multimers have been created by the IntAct database (European Bioinformatics Institute, UK) (e.g. EBI-2325312 for human collagen I), and MatrixDB identifiers are cross-referenced to these complexes. Protein isoforms are identified by a variant number (VARy), and the full MatrixDB identifier becomes MULT_x_VARy_species (e.g. MULT_4_VAR1_human). Matricryptins are identified as PFRAG_x_species and are cross-referenced to the feature identifier of UniProtKB. For example, the MatrixDB identifier of endostatin, a C-terminal fragment of collagen XVIII, is PFRAG_1_human and it is cross-referenced to the UniProtKB feature identifier PRO_0000005794. Glycosaminoglycans (GAG_x), lipids (LIP_x) and cations (CAT_x) are cross-referenced to ChEBI and KEGG compound databases (8, 9). Besides protein–protein and protein–glycosaminoglycan interactions, MatrixDB reports interactions involving cations (mostly calcium) and lipids because a number of extracellular molecules bind to cations and some of them to lipids. Detailed information on each molecule is displayed on the ‘Biomolecule Report Page’.
INTERACTION DATA
MatrixDB is an active member of the International Molecular Exchange (IMEx) consortium (10) and is in charge of the curation of papers published in Matrix Biology, a journal focused on the extracellular matrix, since January 2009. MatrixDB has adopted the PSI-MI standards for annotating and exchanging interaction data. Interaction data stored in MatrixDB are (i) experimentally determined in the laboratory using surface plasmon resonance (SPR) binding assays, including protein and glycosaminoglycan arrays probed by SPR imaging (11), (ii) extracted from the literature by manual curation and (iii) imported from other interaction databases belonging to the IMEx consortium [IntAct (12), DIP (13), MINT (14), BioGRID (15)], as well as from the Human Protein Reference Database (16). Imported data are restricted to interactions involving at least one extracellular protein. The extracellular proteins are identified using UniProtKB/Swiss-Prot keywords and Gene Ontology (17), complemented with manual annotations when required. The text files containing known extracellular human proteins, membrane human proteins and secreted human proteins can be freely downloaded from the download page of MatrixDB. Our curation process has followed the MIMIx guidelines [Minimum Information about a Molecular Interaction experiment; (18)] and has been updated to adhere to the IMEx curation rules in 2010. Interaction data curated by MatrixDB are freely available for download in the PSI-MI XML and TAB 2.5 formats (19).
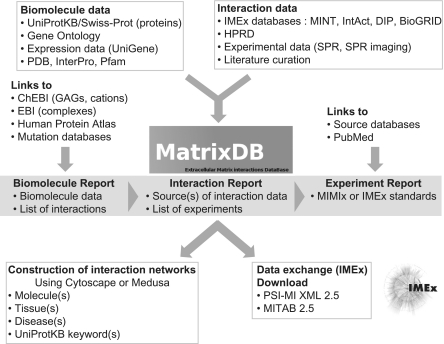
Mammalian interaction data refer to human molecules in order to easily display the list of partners of a given molecule on the ‘Biomolecule Report’ page (cf. the schematic organization of MatrixDB, Figure 1). Clicking on an interaction gives access to the ‘Interaction Report’ page where the source of the data (name of the database) and the experiments supporting the interaction are listed along with links to the abstracts of the corresponding papers. The species experimentally used to demonstrate the interaction are indicated on the ‘Experiment Report’ page with a detailed report of the experiment according to MIMIx or IMEX standards (e.g. interaction detection method, partner detection method, biological and experimental roles of partners, binding sites, kinetics, and affinity when available). MatrixDB is focused on mammalian interactions, but a comprehensive extracellular interaction dataset (69 interactions) of zebrafish has been imported (20,21). We have also curated a recent dataset of the elastic fiber interactome (45 interactions) identified by affinity purification and mass spectrometry (22), and the interactions (∼30) established by SPARC, an extracellular protein involved in a number of biological processes. The current release of MatrixDB contains 2174 extracellular matrix interactions including 1836 protein–protein and 119 protein–glycosaminoglycan interactions. We have curated 490 interactions, and 847 experiments from 192 articles, the other interaction data being imported from several databases (Figure 1). Statistics are available on the ‘Statistics’ page of MatrixDB.
Figure 1.
Organization of MatrixDB showing the sources of biomolecule and interaction data, the ‘Biomolecule Report’, ‘Interaction Report’ and ‘Experiment Report’ pages, the links to other web sites, the construction of interaction networks, data formats available for downloading and data exchange with the members of the IMEx consortium.
INTEGRATION OF LOCALIZATION AND MUTATION DATA
The ‘Biomolecule Report’ page contains a direct link to data from the Human Protein Atlas that shows the expression and localization of proteins in a large variety of normal human tissues, cancer cells and cell lines but is not available for downloading (23). We have imported UniGene expressed sequence tag profiles that reflect approximate expression patterns in tissues [http://www.ncbi.nlm.nih.gov/unigene; (24)] in order to create tissue-specific interaction networks.
We have also added on the Biomolecule Report page a link to databases recapitulating data on mutations occurring in the gene encoding the extracellular protein, including the osteogenesis imperfecta consortium [http://oiprogram.nichd.nih.gov/consortium.html; (25)], a database of osteogenesis imperfecta and Ehlers-Danlos syndrome variants [http://www.le.ac.uk/ge/collagen; (26,27)], and to COLdb, a database linking genetic data to molecular function in fibrillar collagens [http://collagen.stanford.edu/; (28)]. On the ‘Biomolecule Report’ page, and when appropriate, there is a link to the OMIM database of human genes and genetic disorders [http://www.ncbi.nlm.nih.gov/omim; (29)]. These data are used to build disease-specific interaction networks.
MatrixDB: AN EXTRACELLULAR MATRIX WEB SITE
Links to individual extracellular interaction datasets are available on the homepage of MatrixDB. They include the map of candidate cell and matrix interaction domains on the human type I collagen fibril (30), the endostatin interaction network established in our laboratory (11), the elastic fiber interaction network (22) and the cell surface interaction network of neural leucine-rich repeat receptors identified in zebrafish (20,21). Comprehensive extracellular interaction datasets will be curated on a regular basis.
BROWSING MatrixDB
Two types of searches are offered by default. ‘Biomolecule category’ displays all the human molecules in a category (protein, glycosaminoglycan, fragment, lipid, cation and inorganic compound). Searching by ‘Biomolecule name’ can be performed with the biomolecule or gene name or with its UniprotKB and ChEBI accession number or MatrixDB identifier. Three other types of queries are available in the ‘Advanced Search’: free text search, search by PubMed identifier and dataset search. The dataset search displays all the interactions provided by a given database (IntAct, MINT, DIP, BioGRID and MatrixDB), or those reported in specific papers (11,20–22). Detailed data is displayed when a molecule is selected, and links are provided to access further information within MatrixDB or on external websites. For example, UniGene EST profiles or OMIM disease data associated with the gene coding for the protein(s) of interest are provided when available. A list of the protein partners is displayed with the number of experiments reporting each interaction. An interaction can be selected to examine these supporting experiments, and an experiment can be selected to access to kinetics, affinity, binding site and the experimental species.
BUILDING INTERACTION NETWORKS USING MatrixDB DATA
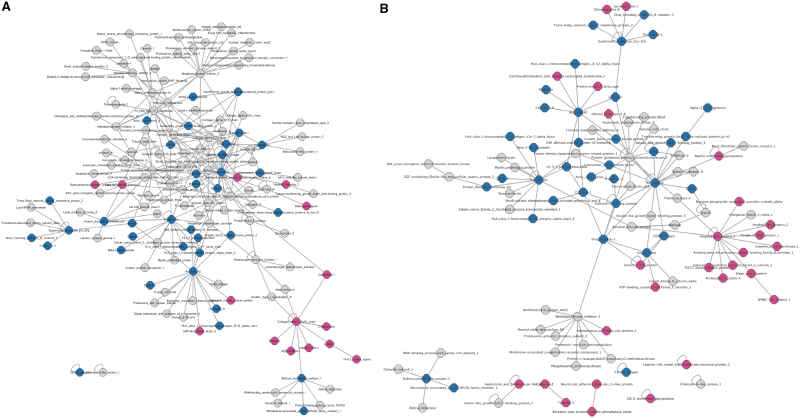
Several options are available for building customized networks. The user can create (i) the entire network of interactions involving at least one extracellular partner, combined or not with interactions established by membrane and secreted molecules, (ii) the interaction network of proteins annotated with user-selected UniProtKB keywords, (iii) the interaction network of one or several molecule(s), including or not the interactions of its (their) partners, (iv) tissue-specific interaction networks (Figure 2) and (v) disease-specific interaction networks. The building of tissue-specific interaction networks is based on expression data imported from UniGene. One or several tissues can be selected and a threshold (minimum number of transcripts per million present in the tissue) can be defined to keep only interactions established by proteins expressed above this threshold in the selected tissues. An option restricts the interactions to those where the partners are specifically expressed in one or several selected tissues. This function allows the identification of tissue-specific partners. It is also possible to build disease-specific interaction networks, based on OMIM identifiers.
Figure 2.
Protein–protein interaction networks of skin (A) and brain (B), built using MatrixDB and visualized with Cytoscape (threshold used for UniGene annotations: ≥100 transcripts per million). Edges: interactions. Pink nodes: proteins specific of skin (A) or brain (B); blue nodes: proteins present in five tissues (liver, lung, bone, skin and brain); grey nodes: other proteins present in two to four tissues out of five.
The interaction networks are visualized using Cytoscape (31) or Medusa (32), using customized styles that are described in the MatrixDB tutorial available on the website.
CONCLUSION
MatrixDB is a database providing interaction data involving extracellular proteins and glycosaminoglycans and interactions established by these two major constituents of the extracellular matrix with cations and lipids. Building the extracellular interactome is a prerequisite to delineate the molecular mechanisms underlying the assembly of the extracellular matrix and to understand how genetic diseases interfere with this process. Future releases will also include interaction data imported from the databases that will join the IMEx consortium. MatrixDB will increase its coverage by curation of interactions involving (i) matrix metalloproteinases and their inhibitors, which play a major role in tissue remodelling (links to the MEROPS database (33) will be provided), (ii) the adhesive matrix molecule family (microbial surface components recognizing adhesive matrix molecules, MSCRAMMs) responsible for the interaction of pathogens with the extracellular matrix (34) and (iii) other proteins and sugars of pathogens.
FUNDING
This work was supported by a CPER grant from the Région Rhône-Alpes; by Institut des Systèmes Complexes (IXXI 2010); and by the EU FP7 ‘PSIMEx’ grant (contract number FP7-HEALTH-2007-223411). Funding for open access charge: EU FP7 ‘PSIMEx’ grant (contract number FP7-HEALTH-2007-223411).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Christophe Blanchet (UMR 5086, Lyon, France) for helping us to install the MatrixDB server, Samuel Kerrien and Bruno Aranda (EBI, Hinxton, UK) for their help regarding data format exchange and Sandra Orchard (EBI, Hinxton, UK) for guiding us through the curation process.
REFERENCES
- 1.Ricard-Blum S, Ruggiero F. The collagen superfamily: from the extracellular matrix to the cell membrane. Pathol. Biol. (Paris) 2005;53:430–442. doi: 10.1016/j.patbio.2004.12.024. [DOI] [PubMed] [Google Scholar]
- 2.Hynes RO. The extracellular matrix: not just pretty fibrils. Science. 2009;326:1216–1219. doi: 10.1126/science.1176009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ricard-Blum S, Ballut L. Matricryptins derived from collagens and proteoglycans. Front Biosci. doi: 10.2741/3712. in press. [DOI] [PubMed] [Google Scholar]
- 4.Bateman JF, Boot-Handford RP, Lamandé SR. Genetic diseases of connective tissues: cellular and extracellular effects of ECM mutations. Nat. Rev. Genet. 2009;10:173–183. doi: 10.1038/nrg2520. [DOI] [PubMed] [Google Scholar]
- 5.Heinegård D. Proteoglycans and more–from molecules to biology. Int. J. Exp. Pathol. 2009;90:575–586. doi: 10.1111/j.1365-2613.2009.00695.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Chautard E, Ballut L, Thierry-Mieg N, Ricard-Blum S. MatrixDB, a database focused on extracellular protein-protein and protein-carbohydrate interactions. Bioinformatics. 2009;2:690–691. doi: 10.1093/bioinformatics/btp025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.de Matos P, Alcantara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Orchard S, Kerrien S, Jones P, Ceol A, Chatr-Aryamontri A, Salwinski L, Nerothin J, Hermjakob H. Submit your interaction data the IMEx way: a step by step guide to trouble-free deposition. Proteomics. 2007;7:28–34. doi: 10.1002/pmic.200700286. [DOI] [PubMed] [Google Scholar]
- 11.Faye C, Chautard E, Olsen BR, Ricard-Blum S. The first draft of the endostatin interaction network. J. Biol. Chem. 2009;284:22041–22047. doi: 10.1074/jbc.M109.002964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ceol A, Chatr AA, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Breitkreutz BJ, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database: 2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Orchard S, Salwinski L, Kerrien S, Montecchi-Palazzi L, Oesterheld M, Stumpflen V, Ceol A, Chatr-Aryamontri A, Armstrong J, Woollard P, et al. The minimum information required for reporting a molecular interaction experiment (MIMIx) Nat. Biotechnol. 2007;25:894–898. doi: 10.1038/nbt1324. [DOI] [PubMed] [Google Scholar]
- 19.Kerrien S, Orchard S, Montecchi-Palazzi L, Aranda B, Quinn AF, Vinod N, Bader GD, Xenarios I, Wojcik J, Sherman D, et al. Broadening the horizon–level 2.5 of the HUPO-PSI format for molecular interactions. BMC Biol. 2007;5:44. doi: 10.1186/1741-7007-5-44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bushell KM, Söllner C, Schuster-Boeckler B, Bateman A, Wright GJ. Large-scale screening for novel low-affinity extracellular protein interactions. Genome Res. 2008;18:622–630. doi: 10.1101/gr.7187808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Söllner C, Wright GJ. A cell surface interaction network of neural leucine-rich repeat receptors. Genome Biol. 2009;10:R99. doi: 10.1186/gb-2009-10-9-r99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Cain SA, McGovern A, Small E, Ward LJ, Baldock C, Shuttleworth A, Kielty CM. Defining elastic fiber interactions by molecular fishing: an affinity purification and mass spectrometry approach. Mol. Cell Proteomics. 2009;8:2715–2732. doi: 10.1074/mcp.M900008-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pontén F, Jirström K, Uhlen M. The human protein atlas–a tool for pathology. J. Pathol. 2008;216:387–393. doi: 10.1002/path.2440. [DOI] [PubMed] [Google Scholar]
- 24.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Marini JC, Forlino A, Cabral WA, Barnes AM, San Antonio JD, Milgrom S, Hyland JC, Körkkö J, Prockop DJ, De Paepe A, et al. Consortium for osteogenesis imperfecta mutations in the helical domain of type I collagen: regions rich in lethal mutations align with collagen binding sites for integrins and proteoglycans. Hum. Mutat. 2007;28:209–221. doi: 10.1002/humu.20429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Dalgleish R. The human type I collagen mutation database. Nucleic Acids Res. 1997;25:181–187. doi: 10.1093/nar/25.1.181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dalgleish R. The human collagen mutation database 1998. Nucleic Acids Res. 1998;26:253–255. doi: 10.1093/nar/26.1.253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bodian DL, Klein TE. COLdb, a database linking genetic data to molecular function in fibrillar collagens. Hum. Mutat. 2009;30:946–951. doi: 10.1002/humu.20978. [DOI] [PubMed] [Google Scholar]
- 29.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick’s Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sweeney SM, Orgel JP, Fertala A, McAuliffe JD, Turner KR, Di Lullo GA, Chen S, Antipova O, Perumal S, Ala-Kokko L, et al. Candidate cell and matrix interaction domains on the collagen fibril, the predominant protein of vertebrates. J. Biol. Chem. 2008;283:21187–21197. doi: 10.1074/jbc.M709319200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Shannon P, Markiel A, Ozier O, Baliga NS, Wang JT, Ramage D, Amin N, Schwikowski B, Ideker T. Cytoscape: a software environment for integrated models of biomolecular interaction networks. Genome Res. 2003;13:2498–2504. doi: 10.1101/gr.1239303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hooper SD, Bork P. Medusa: a simple tool for interaction graph analysis. Bioinformatics. 2005;21:4432–4433. doi: 10.1093/bioinformatics/bti696. [DOI] [PubMed] [Google Scholar]
- 33.Rawlings ND, Barrett AJ, Bateman A. MEROPS: the peptidase database. Nucleic Acids Res. 2010;38:D227–D233. doi: 10.1093/nar/gkp971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Speziale P, Pietrocola G, Rindi S, Provenzano M, Provenza G, Di Poto A, Visai L, Arciola CR. Structural and functional role of Staphylococcus aureus surface components recognizing adhesive matrix molecules of the host. Future Microbiol. 2009;4:1337–1352. doi: 10.2217/fmb.09.102. [DOI] [PubMed] [Google Scholar]