Abstract
Prostate cancer (PC) is one of the most commonly diagnosed cancers in men. PC is relatively difficult to diagnose due to a lack of clear early symptoms. Extensive research of PC has led to the availability of a large amount of data on PC. Several hundred genes are implicated in different stages of PC, which may help in developing diagnostic methods or even cures. In spite of this accumulated information, effective diagnostics and treatments remain evasive. We have developed Dragon Database of Genes associated with Prostate Cancer (DDPC) as an integrated knowledgebase of genes experimentally verified as implicated in PC. DDPC is distinctive from other databases in that (i) it provides pre-compiled biomedical text-mining information on PC, which otherwise require tedious computational analyses, (ii) it integrates data on molecular interactions, pathways, gene ontologies, gene regulation at molecular level, predicted transcription factor binding sites on promoters of PC implicated genes and transcription factors that correspond to these binding sites and (iii) it contains DrugBank data on drugs associated with PC. We believe this resource will serve as a source of useful information for research on PC. DDPC is freely accessible for academic and non-profit users via http://apps.sanbi.ac.za/ddpc/ and http://cbrc.kaust.edu.sa/ddpc/.
BACKGROUND
Prostate cancer (PC) is the second most commonly diagnosed cancer in men and it is the sixth most common cause of cancer death of males. PC constitutes ∼12% of all globally diagnosed male cancers. The rates of incidence and mortality vary according to geographic regions. Within the male population in developed countries PC contributes to 19% of all reported cancer cases and appears to be the most commonly diagnosed male cancer. In developing countries, PC is the sixth most common cancer in men and constitutes ∼5% of all reported male cancer diagnoses (1).
PC is a heterogeneous disease manifesting itself in varying pathological and clinical forms. It is not easy to diagnose and treat as PC tumours may be detected only during autopsy. Significant advancement has been achieved in PC diagnosis with the introduction of prostate-specific antigen (PSA) screening (2). High-throughput genomic technologies such as microarray and gene sequencing have been employed to better understand the molecular mechanisms underlying the development and progression of PC (3). Structural biology and rational drug design, proteomics and cell imaging have also played a major role in understanding receptor and drug interactions in PC (3).
Research in PC has led to the availability of vast amounts of useful data contained in various databases. Prostate expression database (PEDB) accessible at http://www.pedb.org/ is a curated database that contains tools for analysing prostate gene expression in both disease and normal conditions (4). Another database, prostate gene database (PGDB), accessible at http://www.urogene.org/pgdb/ is a curated database on genes or genomic loci related to human prostate and prostatic diseases (5). However, neither of these databases provides insights into the molecular context of gene functioning in PC. Thus, we believe that information on genes experimentally verified as involved in PC enriched with information that provides insights into the molecular context of their functioning in PC, such as information on related proteins, molecular interactions and pathways, drugs and drug targets, transcription regulation, as well as pre-compiled literature-based text-mined reports on PC, will be a useful resource for PC researchers. To the best of our knowledge there is no such web-based resource.
In this study, we present dragon database of genes associated with prostate cancer (DDPC), an integrated knowledge database that has been developed to provide researchers with a multitude of information related to PC and PC-related genes, with the aim to support research of PC at a molecular level.
Description
DDPC contains information on genes that have been experimentally verified as being involved in PC. That information is gathered from very disparate sources. The information is packed in a variety of ways for inspection. For example, DDPC contains pre-compiled text-mined data on PC, gene regulation, ontologies, information on proteins encoded by PC-associated genes, pathways, molecular interactions and drug-related information. Entries in DDPC are linked to external databases such as UniProt (6), SwissProt (7), KEGG pathway database (8), TRANSFAC (9), OMIM (10) and Reactome database (11). Drugs associated with PC have been integrated in DDPC via DrugBank (12). This drug-related information could aid in the search for new cancer drug receptors and potential PC drugs.
Users can query DDPC using keywords such as gene symbols, Entrez Gene IDs or Ensembl Gene IDs or they can set up a batch query. Data can also be retrieved by browsing DDPC using the ‘gene search’, ‘gene select’ and ‘transcription regulation’ catalogues. All data that was retrieved through one of these catalogues can be downloaded as a spreadsheet.
We believe that DDPC, due to its rich information content and rich information on associations of various entities, will be a very useful and novel resource for researchers in the field of PC. DDPC is freely available to academic and non-profit users at http://apps.sanbi.ac.za/ddpc/ and http://cbrc.kaust.edu.sa/ddpc/.
DDPC construction
The DDPC architecture is based on a three-layer structure, namely presentation layer, logic layer and data layer. The logic layer comprises server side PHP and Perl modules, whereas the presentation layer consists of an online graphical user interface developed using DHTML and JavaScript. DDPC is a relational database with a MySQL data layer.
Part of the data in DDPC is manually curated to increase the reliability and relevance of information presented. For curation purposes, a Perl script was used to extract 973 genes from Entrez Gene using the NCBI’s Entrez Programming Utilities (written by Savita Shrivastava, Canadian Bioinformatics Help Desk, University of Alberta, Edmonton) using query strings containing terms such as ‘prostate cancer’ and human(orgn), ‘prostate carcinoma’, ‘prostate adenocarcinoma’ and other PC-related terms. We manually extracted PC-associated genes from that list requesting that a gene’s involvement in PC has experimental support in literature via techniques such as western blot, RT–PCR, immunohistochemistry, tissue microarray, etc. In this way from 973 initially extracted genes only 704 were verified as being involved in PC. Manual curation also provided additional information for each gene [such as PSA levels, Gleason scores (13), clinical and pathological stages of PC], which is incorporated into DDPC to give insights into disease progression. Data statistics for genes incorporated in DDPC are provided at http://apps.sanbi.ac.za/ddpc/statistics.php or http://cbrc.kaust.edu.sa/ddpc/statistics.php in Table 1.
DDPC provides information that may help in understanding of the regulatory potential of PC genes and associated regulatory networks. The database contains information on the genes’ promoters and putative transcription factor binding sites (TFBSs). To generate this information, we extracted 1766 promoters each covering a region of [−1000, +200] relative to the transcription start sites (TSS) of the PC genes. For this we used a FANTOM 3 promoter set based on CAGE libraries (14,15). Mammalian matrix models of TFBSs in TRANSFAC Professional ver. 11.4 (9) were used to map TFBSs to both strands of each promoter. To decrease the incidence of false-positive TFBSs predictions, we used the MatchTM program with a profile for score thresholds that aims to minimize false positives (16). All mammalian transcription factors (TFs) known to be associated with TRANSFAC position weight matrices were used to obtain information on the binding of TFs to promoters of PC genes. A total of 689 mammalian TFs with unique IDs were extracted from TRANSFAC as associated with our TFBS predictions. The information on predicted TFBSs, the associated TFs and the genes they potentially control can be mined in DDPC.
Vital DrugBank data concerning PC genes has been incorporated into DDPC (Table 2 in http://apps.sanbi.ac.za/ddpc/statistics.php or http://cbrc.kaust.edu.sa/ddpc/statistics.php). These comprise ADMET descriptors, metabolizing enzymes, drug targets, coordinate files for proteins and small drug molecules and links to other drug-related databases.
A pre-compiled list of text-mined information from PubMed records has been integrated into DDPC to give the user an overview on potential gene interactions/associations and pathways. To generate the pre-compiled results, the PubMed database was queried using appropriate PC keywords related to each of the PC genes (e.g. ‘A26C3’ OR ‘POTE22’ AND mammal AND cancer) via the NCBI’s Entrez programming utilities. A list of 558 347 abstracts was obtained on 24 February 2009 and query results were analysed using the dragon exploration system (DES) from OrionCell (http://www.orioncell.org), which was based on the ideas outlined in (17,18). DES has been previously used in creation of several other knowledgebases such as DDOC, DDESC and DDEC (19–21). The text-mining dictionaries used by DES in this process were ‘metabolites and enzymes’, ‘genes and proteins’ and ‘chemicals with pharmacological effects’. The text-mining results are shown as lists of tables and graphical representations of interactive networks of genes of interest with other essential biological receptors and pathways.
In order to effectively utilize DDPC, users are encouraged to consult the user manual available at http://apps.sanbi.ac.za/ddpc/manual.php or http://cbrc.kaust.edu.sa/ddpc/manual.php. It can either be viewed in HTML format or downloaded as PDF.
UTILITY AND DISCUSSION
Although existing databases that relate to prostate or PC such as PGDB and PEDB provide a lot of information, they are coming short of providing means to explore the molecular biology of PC. PGDB comprises of information about genes specifically expressed in prostate or documented in literature to be involved in the molecular processes associated with normal or diseased prostate. PGDB provides serial analysis of gene expression (SAGE) or expressed sequence tags (ESTs) expression data for each included gene (5). Another database, PEDB, comprises information of ESTs from prostate cDNA libraries and provides proteomic and experimentally determined microarray data on normal and neoplastic prostate tissues and cell lines (4).
DDPC has been developed not as a substitute for these two repositories, but as a complement to the already available pool of information and resources. DDPC is focused on genes shown experimentally to be involved in PC and it contains information related to these genes and their associations to other concepts of relevance for studies of the molecular background of PC. Much of this information is obtained by novel pre-compiled extensive large-scale analyses of relevant data that otherwise would be very difficult for individual researchers to obtain. Part of the DDPC information is manually curated. Inclusion of genes in DDPC was verified manually to increase the accuracy of the database.
DDPC is a fully searchable web-based knowledgebase. Using the search and browse functionalities, data can be retrieved via ‘gene search’, ‘gene select’, ‘transcription regulation’, ‘batch query’ and ‘drug search’ tabs. For example, clicking on the ‘gene search’ tab enables the user to query the database using the categories ‘Anatomical System’, ‘Cell Line’, ‘KEGG Pathways’ and ‘GO Ontology’. By clicking on any of these categories, users would obtain a list of genes with comprehensive associated data. This data includes the experimental evidence used to identify the gene’s involvement in PC, its related proteins, eVOC ontologies (22), GO ontologies, associated pathways, associated diseases, ortholog genes, transcription regulations, text-mined report and drug-related data, as well as basic information such as gene name, gene symbol, gene ID and links to other resources.
To support studies of the regulatory potential of PC genes and the associated regulatory networks, DDPC provides predicted TFBSs on the promoters of many of the PC genes. This enables the user to understand how the genes may interact with others in relation to their transcription regulatory network. Genes involved in potential pathways may give insight on the invasiveness of cancer cells, which could lead to the understanding of genetic changes occurring during progression (23). These regulatory networks were beneficial in identifying biomarkers for drug discovery, prevention, therapeutic intervention and diagnostic biomarkers for several diseases (24–27).
DDPC also serves as a ‘one-stop shop’ database, where information regarding a particular gene has been extracted from other databases and enriched with information from several additional analyses not obtainable from other repositories. The DDPC incorporated genes were annotated based on eVOC anatomical system category, association with cell type, developmental stage, experimental technique, microarray platform, pathology, taxonomy, tissue preparation and treatment. GO ontologies have also been incorporated into DDPC to help prevent anomalies, which sometimes arise as a result of using multiple terms to describe molecular processes involving genes.
Other essential features of DDPC are the integration of metabolic pathways and text-mining results. Pathway data for genes is mapped in the form of links to OMIM associated diseases as well as to pathway databases that are publicly accessible online without user registration (KEGG and Reactome). KEGG gives information on comprehensively analysed graphical diagrams on known regulatory and metabolic pathways about genes. The Reactome data model enables the integration of mechanisms on intermediary metabolisms, regulative pathways and signal transduction.
In the documentation, we provided instructions how data on a gene can be obtained by browsing the ‘gene select’ catalogue. We used androgen receptor (AR) as an example. By clicking on the AR gene symbol, we retrieved vital comprehensive molecular data on the AR gene in PC development. The involvement of AR in PC was established using immunoreactivity, immunostaining and chromatin immunoprecipitation experiments. Some of the functional annotations obtained from GO ontology were prostate gland development, cell proliferation, cell growth and other insightful concepts. It was established that the gene is involved in the prostate cancer pathway with KEGG entry hsa05215. We obtained further information through OMIM (ID 313700). For example, Newmark et al. (28) proposed that a somatic mutation in the AR gene resulting in persistent expression could lead to androgen-independent PC. Also, some older men with PC have been reported to possess high titers of autoimmune antibodies to androgen receptor (29).
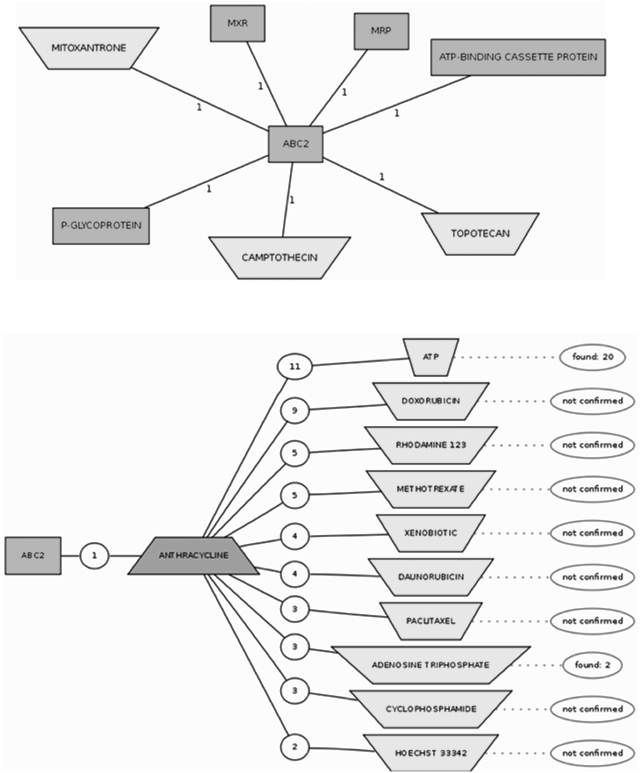
Manual analysis of text is labour intensive. It should be noted that there are more than 500 000 PubMed entries on PC genes. Therefore pre-compiled text-mining reports generated from abstracts of PubMed entries related to PC genes have been incorporated into DDPC. The text-mining reports generate networks as graphical visualizations of essential interactions between different terms. Their representation is easy to understand. The text-mining interactions consist of an interconnected network of colour-coded concepts from various dictionaries with links to the various abstracts containing the concepts. To confirm a relationship retrieved by the system, the user would have to manually verify the abstract in question and either reject or accept the proposed association. The text-mining data also includes a hypothesis generator, which gives information on how different entities may associate with each other. DDPC also contains a list of the most referenced documents (with PubMed IDs) based on the dictionaries used and a table of the various terms contained in the search dictionaries and their frequencies of occurrence in the abstracts. For example, an entity search with the gene ABC2 retrieves a list of colour-coded entities assigned to their respective dictionaries with their frequency of occurrence. This output list could be displayed in a graphical or a table form. These entities co-occur with ABC2 in an abstract and this could infer a possible relationship. Clicking on ‘draw network’ in the menu generates an interaction network of association maps consisting of linked nodes of entities (Figure 1). Also, clicking on ‘generate hypothesis’ retrieves a hypothesis involving ABC2 and other entities. In Figure 1, we have generated hypotheses between ABC2 and other concepts in the metabolites and enzymes dictionary. Verification of the hypotheses revealed that most of the retrieved associations were unconfirmed. In this case, this means that no PubMed abstract reports the co-occurrence of the entities in question.
Figure 1.
Text mining reports in DDPC: an association map between the ABC2 gene and other entity concepts in the form of an interaction network and hypotheses.
Data from DrugBank has also been included in DDPC to complement information about potential drug receptors and drugs. With the incorporation of this data, users can quickly query DDPC to obtain a list of drugs with potential effects to PC. DDPC contains pharmacological, pharmaceutical, structural biology and chemo-informatics data pertaining to these drugs and their receptors. An essential feature of DDPC is the addition of absorption, distribution, metabolism, excretion and toxicity (ADMET) characteristics of a drug molecule, which is critical during the early stages of drug development (30,31). ADMET information has been provided for each drug molecule to aid in the in silico prediction of ADMET descriptors of analogues of these drugs. ADMET descriptors could be used to computationally screen large numbers of drug molecules to eliminate those with possible undesirable ADMET characteristics. The structures of the identified drug molecules could be refined to optimize the ADMET characteristics. One piece of useful data that is provided is ‘percentage protein binding’, which describes the strength of the binding o the drug in question to plasma proteins. This information is essential for the prediction of plasma protein binding percentage for novel candidate drugs. The co-ordinate files of drugs (ligands) provided in DDPC can be downloaded to create ligand libraries for virtual screening or docking to identify lead compounds, and also for drug interaction and metabolism studies. DDPC also contains links to other rich drug knowledgebases such as RxList (http://www.rxlist.com/script/main/hp.asp) (32), KEGG drug (http://www.genome.jp/kegg/drug/) (8), KEGG compound (http://www.genome.jp/kegg/compound/) (8), PubChem compound (http://www.ncbi.nlm.nih.gov/pccompound) (33), PubChem substance (http://www.ncbi.nlm.nih.gov/pcsubstance) (33) and PharmGKB (http://www.pharmgkb.org/) (34).
CONCLUSION
We have developed a PC specific knowledgebase consisting of information on genes reported to be implicated in PC. This is a comprehensive database that integrates a variety of data from other biological sources. The genes included in DDPC are also cross-referenced to external databases as leverage for harnessing new biological insights. The incorporation of DrugBank drugs and drug targets associated with PC could potentially help biologist to relate molecular data to pharmacokinetics of drugs. The incorporation of post-processed textual data on PC-related MEDLINE abstracts downloaded via PubMed could be useful for identification of new hypothesis and potential associations between disease concepts and cellular drug targets. This database is a repository that provides essential information for researchers working on PC and is aimed at facilitating integration of cross-discipline resources to accelerate answering pertinent biological questions involving PC.
FUTURE DEVELOPMENTS
We plan to expand DDPC both in content and functionality. DDPC will be updated annually. New data on protein–protein interactions and microarrays expression experiments will be integrated. The gene lists will be expanded and the present PC genes will be checked for being up-to-date. All text mining reports will be re-compiled with up-to-date information from PubMed and drug data will be updated based on the current information available in DrugBank. Incorporating additional search and retrieval features will enhance the usability of the web interface. Although a number of databases for information regarding PC exist, DDPC is a unique resource because it unifies three distinctive characteristics in a single application. It provides pre-compiled biomedical text-mining information on PC. It integrates data on molecular interactions, pathways, gene ontologies, gene regulation at molecular level based on predicted transcription factor binding sites on promoters of PC implicated genes and transcription factors that correspond to these binding sites. And it contains DrugBank data on drugs associated with PC.
FUNDING
The National Research Foundation of South Africa (to M.M.); The National Bioinformatics Network grants (partial); Centre for Disease Control (to E.O.), Atlanta, Georgia, US Government; The National Research Chair grant (to A.C.); S.K.K. was partially supported by the grants from the National Bioinformatics Network (61070 to V.B.B.), DST/NRF Research Chair (64751 to V.B.B.) and National Research Foundation (62302 to V.B.B.); While in South Africa A.R., U.S. and V.B.B. were partially supported by the grants from the National Bioinformatics Network (61070 to V.B.B.), DST/NRF Research Chair (64751 to V.B.B.) and National Research Foundation (62302 to V.B.B.); M.K. has been supported by a postdoctoral fellowship from the Claude Leon Foundation, South Africa. Funding for open access charge: V.B. KAUST research funds.
Conflict of interest statement. V.B.B. and A.R. are partners in the OrionCell Company, whose product dragon exploration system (DES) is used in creation of DDPC pre-compiled reports. Other contributing authors declare no conflict of interest.
ACKNOWLEDGEMENT
M.M., M.K. and V.B.B. conceptualized the study, analysed results and wrote the manuscript. S.K., A.R., U.S., S.S., E.K. and A.C. contributed to different aspects of the analysis and data preparation. A.R. and V.B.B. developed the DES system.
RERERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global Cancer Statistics, 2002. CA Cancer J. Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.Wang MC, Valenzuela LA, Murphy GP, Chu TM. Purification of a human prostate specific antigen. Invest. Urol. 1979;17:159–163. [PubMed] [Google Scholar]
- 3.Reynolds MA. Molecular alterations in prostate cancer. Cancer Lett. 2008;271:13–24. doi: 10.1016/j.canlet.2008.04.047. [DOI] [PubMed] [Google Scholar]
- 4.Hawkins V, Doll D, Bumgarner R, Smith T, Abajian C, Hood L, Nelson PS. PEDB: the Prostate Expression Database. Nucleic Acids Res. 1999;27:204–208. doi: 10.1093/nar/27.1.204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li L, Zhao H, Shiina H, Kane CJ, Dahiya R. PGDB: a curated and integrated database of genes related to the prostate. Nucleic Acids Res. 2003;31:291–293. doi: 10.1093/nar/gkg008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gasteiger E, Jung E, Bairoch A. SWISS-PROT: connecting biomolecular knowledge via a protein database. Curr. Issues Mol. Biol. 2001;3:47–55. [PubMed] [Google Scholar]
- 8.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC and its module TRANSCompel: transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gleason DF, Mellinger GT. Prediction of prognosis for prostatic adenocarcinoma by combined histological grading and clinical staging. J. Urol. 1974;111:58–64. doi: 10.1016/s0022-5347(17)59889-4. [DOI] [PubMed] [Google Scholar]
- 14.Carninci P, Kasukawa T, Katayama S, Gough J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al. The transcriptional landscape of the mammalian genome. Science. 2005;309:1559–1563. doi: 10.1126/science.1112014. [DOI] [PubMed] [Google Scholar]
- 15.Carninci P, Sandelin A, Lenhard B, Katayama S, Shimokawa K, Ponjavic J, Semple CAM, Taylor MS, Engström PG, Frith MC, et al. Genome-wide analysis of mammalian promoter architecture and evolution. Nat. Genet. 2006;38:626–635. doi: 10.1038/ng1789. [DOI] [PubMed] [Google Scholar]
- 16.Kel AE, Gössling E, Reuter I, Cheremushkin E, Kel-Margoulis OV, Wingender E. MATCH: a tool for searching transcription factor binding sites in DNA sequences. Nucleic Acids Res. 2003;31:3576–3579. doi: 10.1093/nar/gkg585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Pan H, Zuo L, Choudhary V, Zhang Z, Leow SH, Chong FT, Huang Y, Ong VWS, Mohanty B, Tan SL, et al. Dragon TF Association Miner: a system for exploring transcription factor associations through text-mining. Nucleic Acids Res. 2004;32:W230–W234. doi: 10.1093/nar/gkh484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bajic VB, Veronika M, Veladandi PS, Meka A, Heng M, Rajaraman K, Pan H, Swarup S. Dragon Plant Biology Explorer. A text-mining tool for integrating associations between genetic and biochemical entities with genome annotation and biochemical terms lists. Plant Physiol. 2005;138:1914–1925. doi: 10.1104/pp.105.060863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kaur M, Radovanovic A, Essack M, Schaefer U, Maqungo M, Kibler T, Schmeier S, Christoffels A, Narasimhan K, Choolani M, et al. Database for exploration of functional context of genes implicated in ovarian cancer. Nucleic Acids Res. 2009;37:D820–D823. doi: 10.1093/nar/gkn593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sagar S, Kaur M, Dawe A, Seshadri SV, Christoffels A, Schaefer U, Radovanovic A, Bajic VB. DDESC: Dragon database for exploration of sodium channels in human. BMC Genomics. 2008;9:622. doi: 10.1186/1471-2164-9-622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Essack M, Radovanovic A, Schaefer U, Schmeier S, Seshadri SV, Christoffels A, Kaur M, Bajic VB. DDEC: Dragon database of genes implicated in esophageal cancer. BMC Cancer. 2009;9:219. doi: 10.1186/1471-2407-9-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kelso J, Visagie J, Theiler G, Christoffels A, Bardien S, Smedley D, Otgaar D, Greyling G, Jongeneel CV, McCarthy MI, et al. eVOC: a controlled vocabulary for unifying gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Anastassiou D. Computational analysis of the synergy among multiple interacting genes. Mol. Syst. Biol. 2007;3:83. doi: 10.1038/msb4100124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H, Zhan M. Systematic intervention of transcription for identifying network response to disease and cellular phenotypes. Bioinformatics. 2006;22:96–102. doi: 10.1093/bioinformatics/bti752. [DOI] [PubMed] [Google Scholar]
- 25.Shmulevich I, Dougherty ER, Zhang W. Gene perturbation and intervention in probabilistic Boolean networks. Bioinformatics. 2002;18:1319–1331. doi: 10.1093/bioinformatics/18.10.1319. [DOI] [PubMed] [Google Scholar]
- 26.Pal R, Datta A, Bittner ML, Dougherty ER. Intervention in context-sensitive probabilistic Boolean networks. Bioinformatics. 2005;21:1211–1218. doi: 10.1093/bioinformatics/bti131. [DOI] [PubMed] [Google Scholar]
- 27.Kothandaraman N, Bajic VB, Brendan PNK, Huak CY, Keow PB, Razvi K, Salto-Tellez M, Choolani M. E2F5 status significantly improves malignancy diagnosis of epithelial ovarian cancer. BMC Cancer. 2010;10:64. doi: 10.1186/1471-2407-10-64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newmark JR, Hardy DO, Tonb DC, Carter BS, Epstein JI, Isaacs WB, Brown TR, Barrack ER. Androgen receptor gene mutations in human prostate cancer. Proc. Natl Acad. Sci. USA. 1992;89:6319–6323. doi: 10.1073/pnas.89.14.6319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liao S, Witte D. Autoimmune anti-androgen-receptor antibodies in human serum. Proc. Natl Acad. Sci. USA. 1985;82:8345–8348. doi: 10.1073/pnas.82.24.8345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jónsdóttir SO, Jørgensen FS, Brunak S. Prediction methods and databases within chemoinformatics: emphasis on drugs and drug candidates. Bioinformatics. 2005;21:2145–2160. doi: 10.1093/bioinformatics/bti314. [DOI] [PubMed] [Google Scholar]
- 31.Tsaioun K, Bottlaender M, Mabondzo A. ADDME–Avoiding Drug Development Mistakes Early: central nervous system drug discovery perspective. BMC Neurol. 2009;9(Suppl. 1):S1. doi: 10.1186/1471-2377-9-S1-S1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Hatfield CL, May SK, Markoff JS. Quality of consumer drug information provided by four Web sites. Am. J. Health Syst. Pharm. 1999;56:2308–2311. doi: 10.1093/ajhp/56.22.2308. [DOI] [PubMed] [Google Scholar]
- 33.Sayers EW, Barrett T, Benson DA, Bolton E, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2010;38:D5–D16. doi: 10.1093/nar/gkp967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hodge AE, Altman RB, Klein TE. The PharmGKB: integration, aggregation, and annotation of pharmacogenomic data and knowledge. Clin. Pharmacol. Ther. 2007;81:21–24. doi: 10.1038/sj.clpt.6100048. [DOI] [PubMed] [Google Scholar]