Abstract
FlyFactorSurvey (http://pgfe.umassmed.edu/TFDBS/) is a database of DNA binding specificities for Drosophila transcription factors (TFs) primarily determined using the bacterial one-hybrid system. The database provides community access to over 400 recognition motifs and position weight matrices for over 200 TFs, including many unpublished motifs. Search tools and flat file downloads are provided to retrieve binding site information (as sequences, matrices and sequence logos) for individual TFs, groups of TFs or for all TFs with characterized binding specificities. Linked analysis tools allow users to identify motifs within our database that share similarity to a query matrix or to view the distribution of occurrences of an individual motif throughout the Drosophila genome. Together, this database and its associated tools provide computational and experimental biologists with resources to predict interactions between Drosophila TFs and target cis-regulatory sequences.
INTRODUCTION
The first critical step in converting genomic sequence into temporally and spatially patterned gene expression is the regulation of transcription. This process is typically controlled by cis-regulatory modules (CRMs), discrete sequences that contain groups of binding sites for sequence specific transcription factors (TFs). Experimental methods such as chromatin immunoprecipitation allow direct genome-wide analysis of TF binding in a specific cell type and experimental condition (1–4). Work in Drosophila and other organisms has shown that matrix representations of the recognition motif of a TF can be used to computationally map enrichment of its binding sites across the genome (5–7). By analyzing homo- and heterotypic clusters of TF binding sites, conservation of these sites across species and the spatial and temporal expression of TFs and their potential target genes, it is possible to computationally construct transcription regulatory networks (6,8–10). In the case of the Drosophila anterior–posterior patterning network, we have shown that the accuracy of networks predicted based on a nearly complete set of TF binding site motifs can be similar to that obtained using chromatin immunoprecipitation data for these TFs (11). In principle, such computational approaches could be applied in any cell type where sufficiently complete gene expression and TF binding specificity data is available. Currently, one major limitation for this type of analysis is the incomplete description of recognition motifs for the majority of sequence-specific TFs.
The first studies of TF DNA binding specificity used biochemical methods such as DNase I footprinting to identify individual binding sites in known target regulatory sequences. Compilations of these sites (12,13) have provided a rich but crude source of descriptions of binding site preferences. In Drosophila, motifs constructed from these compiled sites (14) provided a basis for many early studies of TF–CRM regulatory interactions. Subsequently, a variety of additional methods have been developed to study binding specificities more systematically, including systematic evolution of ligands by exponential enrichment (SELEX) (15–17), SELEX with deep sequencing (18–20) and protein binding microarrays (PBMs) (21). As an alternative to these purely in vitro methods, we have developed the bacterial one-hybrid system (B1H) that allows TF specificities to be determined without purification and in the context of competition from the bacterial genome (22). The relative efficiency of this system has allowed for the systematic characterization of large numbers of TFs in Drosophila, including all members of the large family of homeodomain proteins and all of the known core components of the embryonic A-P patterning network (23,24). This method allows relatively large libraries of randomized binding sites (∼108) to be rapidly interrogated for potential recognition sequences, where hundreds to thousands of binding sites can be recovered and characterized using high-throughput SOLEXA sequencing.
Several existing databases house collections of TF DNA binding information. The commercial database Transfac (12) and the publically accessible database JASPAR (13) both include matrix descriptions of recognition motifs for TFs across multiple species generated from a variety of methodologies, including compiled sequences, SELEX, PBMs and B1H (13,25,26). The Redfly database, which is specific to Drosophila, provides an extensive compilation of published experimental data identifying CRMs and individual TF binding sites within these CRMs (27). The Uniprobe database provides specificity information for TFs derived from a single technique, PBMs, providing access to the underlying raw data, which allows investigators to directly employ the binding site preferences determined by the data producer or to develop alternative representations of these data to describe recognition (28).
FlyFactorSurvey (FFS) provides an important complement to these existing databases. The current version focuses exclusively on the description of DNA binding specificities for Drosophila TFs determined by B1H and other methods. This database provides a repository for an ongoing project to determine specificities for all TFs in Drosophila, which is one of the primary model organisms for the analysis of transcriptional regulatory networks in metazoa. This database houses more than 400 recognition motifs for over 200 factors often generated from hundreds to thousands of selected binding sites. In keeping with the spirit of other genome-wide analysis projects, a large number of these binding specificities have been released prior to publication to facilitate the use of this information for the analysis of transcriptional regulation at the level of individual TFs, genes or regulatory pathways.
DATABASE CONTENT
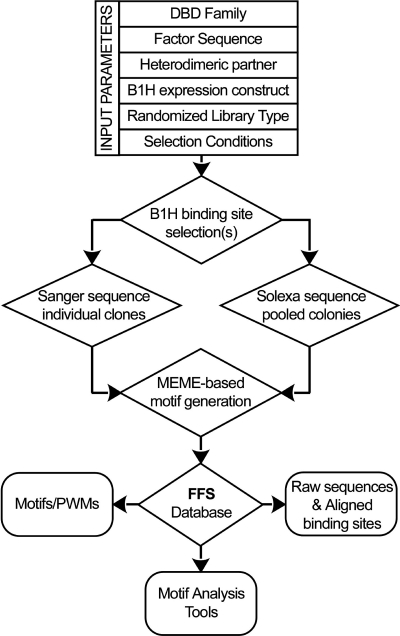
The primary source of recognition motifs within FlyFactorSurvey is TF binding site selections performed using the B1H method (22–24). An outline of the important selection parameters captured in the database, as well as the data processing pipeline is shown in Figure 1. In brief, the predicted DNA binding domain of each Drosophila TF is expressed as a fusion to a component of Escherichia coli RNA polymerase and transformed into cells with a library of reporter plasmids containing a randomized DNA sequence upstream of a weak promoter driving expression of the His3 gene. When plated on media lacking histidine and containing a His3 inhibitor, plasmids with a complementary binding site within the randomized region are required for colony growth. TF binding sites are recovered by sequencing the randomized region of the reporter plasmid recovered from visible colonies. The MEME algorithm (29,30) is used to identify enriched sequence motifs within these sequences, which should represent the DNA binding specificity of the assayed TF. These motifs can be represented in a variety of formats, including alignments of the sequences containing the motifs, a counts matrix depicting the number of sequences with a given base at each position and a position weight matrix (PWM) depicting the log-odds score at each position (31). FlyFactorSurvey houses three classes of information regarding each selection. First, some general information about each TF is provided, including direct links to other wide-ranging databases describing Drosophila genes and proteins, such as FlyBase and FlyMine (32,33). Second, parameters of B1H selections and sequence analysis are described. Third, recovered sequences, the sequence motif output from MEME and images depicting the information content in these motifs are stored.
Figure 1.
Schematic of data flow into the FlyFactorSurvey (FFS) database. The majority of motifs present in the database originate from B1H binding site selections. Information on the factor constructs and selection conditions is captured within the database for each motif generated. Clones from each selection are sequenced either individually via Sanger sequencing or as a pooled population via SOLEXA sequencing, and binding site motifs are identified from these sequences using MEME (39). The FFS database provides users with access to published and unpublished motif information on our characterized Drosophila TFs as well as links to tools to mine our database of motifs and utilize these motifs for searches within the Drosophila genome.
Within the B1H system a variety of parameters can be adjusted to ensure a successful selection or change the complexity of the recovered recognition motif. Many of these important parameters are captured with the database (Figure 1). For each TF characterized using the B1H system, the type of the DNA-binding domain(s) and the amino acid sequence of the portion of the protein characterized is provided. In some cases this is the entire coding sequence, but in others it is the region spanning the DNA-binding domain as well as conserved flanking elements that may play accessory roles in recognition. For heterodimeric TF complexes characterized by the B1H method, the amino acid sequence of both protein fragments is included. The TF expression vector employed dictates the TF fusion partner (either the alpha or omega subunit of E. coli RNA polymerase) and the strength of the promoter (lppC > UV5 > UV2) driving expression of the hybrid protein. In addition, some vectors contain two zinc fingers from Zif268 (‘zif12’) as an accessory DNA-binding domain to increase the activity of the hybrid protein (23). Finally, a subset of these vectors allows expression of a second monomer as an untethered protein for studies of heterodimeric complexes. The vector name provided within the database captures each of these parameters. Additional selection parameters captured in the database include the type of binding site library, which can vary in the length of the randomized region and can contain a fixed DNA binding site for the zif12 protein fusion partner. The stringency of the selection is primarily influenced by the concentration of the inducer Isopropyl β-d-1-thiogalactopyran (IPTG) regulating TF expression and concentration of a His3 inhibitor (3AT), where higher concentrations of inhibitor require higher affinity interactions between the hybrid protein and DNA binding site for colony growth.
TF DNA binding sites are characterized from bacterial colonies by two methods. First, the randomized region is amplified and sequenced (Sanger method) from 24 to 48 individual colonies. MEME is used to identify an overrepresented sequence motif within this population of sequences that should represent the recognition motif of the TF. If a significant motif is identified, the randomized region is amplified from a pool of all colonies on the selection plate, characterized by SOLEXA sequencing and the unique sequences are analyzed using MEME. The Sanger-generated motif is compared with the SOLEXA-generated motif as a quality control step. Aligned sequences comprising the MEME-identified motif are entered into WebLogo (34) to generate a “Sequence logo” (35) describing the information content at each position in the recovered motif. For each recognition motif, the associated unique sequences, aligned sequences, counts matrix and WebLogo image are captured within the database. For published B1H motifs, the associated Pubmed ID number is also provided.
FlyFactorSurvey can also host DNA binding specificity information derived from other experimental methods. In these cases, the construct and selection information may be incomplete, but counts matrices and WebLogo images describing the motif can still be generated and associated with a given TF and publication. The database currently contains this information for DNase I footprint-derived motifs described by Bergman et al. (14).
DATABASE STRUCTURE AND USER INTERFACE
Database schema and website
The FFS database application was developed in house using a MySQL relational database hosted in a database server as the back-end, with the business/presentation layer written using PHP and client access through a standard web client (browser) hosted by a Apache server. The database consists of several tables. The TF table stores detailed information about the TFs. The PWM table stores information about the PWMs. The TF_PWM table links TFs to PWMs allowing a many-to-many relationship. The DNA_BindingSites table contains the raw selected sequences as well as the aligned sequences used to derive the PWM. The selection table contains the detailed selection conditions used to obtain the sequences that ultimately generated the PWMs. The DNAbindingDomain table contains detailed information about the DNA binding domains associated with the TFs. Lastly, the users table stores user information and associated roles for access control and monitoring. The database contains constraints, indices and keys to ensure data integrity and high performance. In addition, each editable record contains meta-data to monitor recent edits with regards to the user, time and location for any changes. The detailed relationship among these tables can be viewed as the entity relation diagram (ERD) at http://pgfe.umassmed.edu/TFDBS/Documentation/FFS_schema.pdf.
Data access
The website home page provides several paths to navigate to TF binding site data of interest. The header contains a link to a ‘browse’ page that lists all TFs with associated DNA binding specificity data within the database (currently 250 factors). Links to individual TF summary pages—one per factor—are on the left of each table in the ‘view’ column. The home page also provides two search windows that allow users to either search for TFs of interest based on gene name or other identification information, or to search for TFs based on the presence of the type of DNA binding domain motif contained within the protein (e.g. homeodomain or C2H2 zinc finger). Each of these searches can be restricted to comprise only TFs with associated binding specificity information within the database or only TFs characterized using the B1H method. These searches return matching TFs in the same format provided in the browse page.
The TF summary page for each factor is split into two sections (Figure 2). The top section provides key descriptive information for the TF, including links to the FlyBase (32,36) and FlyMine (33) databases. Below the TF summary are individual panels for each associated recognition motif. In most cases, multiple motifs are available for each TF. These may be derived from different methods to identify binding sites (B1H and DNase I), different sequencing methods, different selection conditions or different expression constructs. In addition, TFs that recognize their binding sites as dimers may have different motifs associated with different hetero- or homodimeric binding partners. For each recognition motif, the WebLogo image is shown and download buttons allow retrieval of the aligned binding sites, all unique raw sequences analyzed to discover the motif or matrices representing the motif in formats compatible with different sequence analysis programs. Information describing the parameters of the B1H selection that generated the motif is provided on the right side of each motif. In the case of selections performed with heterodimers, a link to the TF summary page for the partner and its amino acid sequence are also displayed.
Figure 2.
Screen shot of bicoid summary page within FlyFactorSurvey. (top) Header information for each factor contains identification information, the type(s) of DNA binding domains found within the gene and links to factor information in Flybase (36), Unipro (40) and FlyMine (33). (bottom) Recognition motifs determined for the factor through different methods, under different conditions or assayed via different sequencing methods are displayed in independent panels. Each panel displays the recognition motif as a Sequence logo (35) and download buttons to obtain count, position-specific probability (PSPM) or position-specific scoring (PSSM) matrices. The other information panel summarizes the methods used to select and sequence the factor binding sites. For motifs determined using the B1H system, the expression vector and the selection conditions are indicated where different stringencies can result in motifs with different complexity. In this example, only two of the four bicoid motif panels within the database are shown; these panels illustrate that the increased inhibitor concentration used in the selection to generate the lower panel resulted in a more stringent motif. For each individual motif, a direct link is provided to view the relative frequency of the motif in the Drosophila genome using Genome Surveyor (see Figure 3).
FlyFactorSurvey also provides an option for obtaining binding site information and matrices for all stored motifs as a flat file download. The home page header contains a link to a Downloads and Resources page. As on each TF summary page, the motif sequences and matrices for the entire dataset are accessible in a variety of formats to maximize the ability of biologists to employ data for large numbers of factors with existing computational tools.
Analysis tools
The sequence and motif formats provided for download are compatible with many commonly used computational tools to analyze TF binding specificities and to map potential binding sites for a TF or set of TFs within a genomic sequence. Two analysis tools of use to the Drosophila research community have been integrated with this database. The first is an implementation of the TOMTOM motif comparison tool from the MEME suite (29,37). A version of the tool populated with all FlyFactorSurvey motifs is accessible via a link from the home page. This program allows a user to input a query motif and identify similar motifs within the database. For example, an investigator might identify an enriched motif in promoters of genes expressed in a given cell type and then query whether any of the TFs in FlyFactorSurvey have a similar DNA binding specificity. Each resulting match provides an image of the query and subject motifs and a link to the TF summary page containing the matching motif within the database. An example of such a search and the resulting TOMTOM output is described in Supplementary Figure S1.
The second tool is an implementation of GenomeSurveyor that allows investigators to examine the distribution of matches to a given DNA binding site motif within regions of the Drosophila genome. GenomeSurveyor uses a hidden Markov model to score DNA binding site motif matches for single or multiple TFs in 500-bp regions of the Drosophila genome and then display them as Z-score tracks on a genome browser (Gbrowse) (7,24,38). Within the TF summary page for a TF in FlyFactorSurvey, each listed motif has a link to a GenomeSurveyor page where the default settings display three Z-score tracks for the motif calculated over the Drosophila melanogaster genome and as averages across the genomes of two or eleven Drosophila species (Figure 3). An additional track displays the position of individual high scoring matches to the motif. When viewed in smaller genomic regions, the matching genomic sequence can be directly observed. The default genomic region (20 kB surrounding the eve gene) can be shifted to any desired region of the genome.
Figure 3.
Screen shot of Genome Surveyor interface directly linked from the Bcd_SOLEXA motif within the FFS database. The relative enrichment (profile) of this motif in 500 bp windows surrounding the eve locus is represented as a Z score relative to the genome-wide average. Three different motif profiles are shown: Single species (D. melanogaster), Multi species (D. melanogaster and D. pseudoobscura) and BMPhylo (11 Drosophila species). BMPhylo motif tracks have been previously described (11). Individual high scoring sequence motif matches are also shown at bottom. This tool provides a rapid assessment of the overrepresentation of any motif in the database within the Drosophila genome and additional functions such as combined motif searches (24).
AVAILABILITY
All data is freely available for distribution at the website. The authors request that they be contacted if multiple unpublished motifs from the database are being used prior to formal publication by the authors. Database and website code are available on request.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The characterization and analysis of transcription factor binding specificities, for the construction of the database and website. Funding for open access charge: National Human Genome Research Institute of the National Institutes of Health (R01 HG004744-01 to M.H.B. and S.A.W.).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Marcus Noyes and Adam Richards for their contributions to the initial stages of this project.
REFERENCES
- 1.Johnson DS, Mortazavi A, Myers RM, Wold B. Genome-Wide Mapping of in Vivo Protein-DNA Interactions. Science. 2007;316:1497–1502. doi: 10.1126/science.1141319. [DOI] [PubMed] [Google Scholar]
- 2.Li XY, Macarthur S, Bourgon R, Nix D, Pollard DA, Iyer VN, Hechmer A, Simirenko L, Stapleton M, Hendriks CL, et al. Transcription Factors Bind Thousands of Active and Inactive Regions in the Drosophila Blastoderm. PLoS Biol. 2008;6:e27. doi: 10.1371/journal.pbio.0060027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ren B, Robert F, Wyrick JJ, Aparicio O, Jennings EG, Simon I, Zeitlinger J, Schreiber J, Hannett N, Kanin E, et al. Genome-wide location and function of DNA binding proteins. Science. 2000;290:2306–2309. doi: 10.1126/science.290.5500.2306. [DOI] [PubMed] [Google Scholar]
- 4.Zeitlinger J, Zinzen RP, Stark A, Kellis M, Zhang H, Young RA, Levine M. Whole-genome ChIP-chip analysis of Dorsal, Twist, and Snail suggests integration of diverse patterning processes in the Drosophila embryo. Genes Dev. 2007;21:385–390. doi: 10.1101/gad.1509607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Berman BP, Pfeiffer BD, Laverty TR, Salzberg SL, Rubin GM, Eisen MB, Celniker SE. Computational identification of developmental enhancers: conservation and function of transcription factor binding-site clusters in Drosophila melanogaster and Drosophila pseudoobscura. Genome Biol. 2004;5:R61. doi: 10.1186/gb-2004-5-9-r61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Schroeder MD, Pearce M, Fak J, Fan H, Unnerstall U, Emberly E, Rajewsky N, Siggia ED, Gaul U. Transcriptional control in the segmentation gene network of Drosophila. PLoS Biol. 2004;2:E271. doi: 10.1371/journal.pbio.0020271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Sinha S, Schroeder MD, Unnerstall U, Gaul U, Siggia ED. Cross-species comparison significantly improves genome-wide prediction of cis-regulatory modules in Drosophila. BMC Bioinformatics. 2004;5:129. doi: 10.1186/1471-2105-5-129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Segal E, Raveh-Sadka T, Schroeder M, Unnerstall U, Gaul U. Predicting expression patterns from regulatory sequence in Drosophila segmentation. Nature. 2008;451:535–540. doi: 10.1038/nature06496. [DOI] [PubMed] [Google Scholar]
- 9.Kheradpour P, Stark A, Roy S, Kellis M. Reliable prediction of regulator targets using 12 Drosophila genomes. Genome Res. 2007;17:1919–1931. doi: 10.1101/gr.7090407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Janssens H, Hou S, Jaeger J, Kim AR, Myasnikova E, Sharp D, Reinitz J. Quantitative and predictive model of transcriptional control of the Drosophila melanogaster even skipped gene. Nat. Genet. 2006;38:1159–1165. doi: 10.1038/ng1886. [DOI] [PubMed] [Google Scholar]
- 11.Blatti C, Richards A, McCutchan M, Wakabayashi-Ito N, Hammonds AS, Celniker SE, Kumar S, Wolfe SA, Brodsky MH, Sinha S. Quantitative analysis of the Drosophila segmentation regulatory network using pattern generating potentials. PLoS Biol. 2010;8 doi: 10.1371/journal.pbio.1000456. PMID: 20808951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matys V, Kel-Margoulis OV, Fricke E, Liebich I, Land S, Barre-Dirrie A, Reuter I, Chekmenev D, Krull M, Hornischer K, et al. TRANSFAC(R) and its module TRANSCompel(R): transcriptional gene regulation in eukaryotes. Nucleic Acids Res. 2006;34:D108–D110. doi: 10.1093/nar/gkj143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Portales-Casamar E, Thongjuea S, Kwon AT, Arenillas D, Zhao X, Valen E, Yusuf D, Lenhard B, Wasserman WW, Sandelin A. JASPAR 2010: the greatly expanded open-access database of transcription factor binding profiles. Nucleic Acids Res. 2010;38:D105–D110. doi: 10.1093/nar/gkp950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bergman CM, Carlson JW, Celniker SE. Drosophila DNase I footprint database: a systematic genome annotation of transcription factor binding sites in the fruitfly, Drosophila melanogaster. Bioinformatics. 2005;21:1747–1749. doi: 10.1093/bioinformatics/bti173. [DOI] [PubMed] [Google Scholar]
- 15.Kinzler KW, Vogelstein B. The GLI gene encodes a nuclear protein which binds specific sequences in the human genome. Mol. Cell Biol. 1990;10:634–642. doi: 10.1128/mcb.10.2.634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tuerk C, Gold L. Systematic evolution of ligands by exponential enrichment: RNA ligands to bacteriophage T4 DNA polymerase. Science. 1990;249:505–510. doi: 10.1126/science.2200121. [DOI] [PubMed] [Google Scholar]
- 17.Roulet E, Busso S, Camargo AA, Simpson AJ, Mermod N, Bucher P. High-throughput SELEX-SAGE method for quantitative modeling of transcription-factor binding sites. Nat. Biotechnol. 2002;20:831–835. doi: 10.1038/nbt718. [DOI] [PubMed] [Google Scholar]
- 18.Zhao Y, Granas D, Stormo GD. Inferring binding energies from selected binding sites. PLoS Comput. Biol. 2009;5:e1000590. doi: 10.1371/journal.pcbi.1000590. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zykovich A, Korf I, Segal DJ. Bind-n-Seq: high-throughput analysis of in vitro protein-DNA interactions using massively parallel sequencing. Nucleic Acids Res. 2009;37:e151. doi: 10.1093/nar/gkp802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jolma A, Kivioja T, Toivonen J, Cheng L, Wei G, Enge M, Taipale M, Vaquerizas JM, Yan J, Sillanpaa MJ, et al. Multiplexed massively parallel SELEX for characterization of human transcription factor binding specificities. Genome Res. 2010;20:861–873. doi: 10.1101/gr.100552.109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Berger MF, Bulyk ML. Universal protein-binding microarrays for the comprehensive characterization of the DNA-binding specificities of transcription factors. Nat. Protoc. 2009;4:393–411. doi: 10.1038/nprot.2008.195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Meng X, Brodsky MH, Wolfe SA. A bacterial one-hybrid system for determining the DNA-binding specificity of transcription factors. Nat. Biotechnol. 2005;23:988–994. doi: 10.1038/nbt1120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Noyes MB, Christensen RG, Wakabayashi A, Stormo GD, Brodsky MH, Wolfe SA. Analysis of homeodomain specificities allows the family-wide prediction of preferred recognition sites. Cell. 2008;133:1277–1289. doi: 10.1016/j.cell.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Noyes MB, Meng X, Wakabayashi A, Sinha S, Brodsky MH, Wolfe SA. A systematic characterization of factors that regulate Drosophila segmentation via a bacterial one-hybrid system. Nucleic Acids Res. 2008;36:2547–2560. doi: 10.1093/nar/gkn048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wingender E. The TRANSFAC project as an example of framework technology that supports the analysis of genomic regulation. Brief Bioinform. 2008;9:326–332. doi: 10.1093/bib/bbn016. [DOI] [PubMed] [Google Scholar]
- 26.Wingender E, Chen X, Hehl R, Karas H, Liebich I, Matys V, Meinhardt T, Pruss M, Reuter I, Schacherer F. TRANSFAC: an integrated system for gene expression regulation. Nucleic Acids Res. 2000;28:316–319. doi: 10.1093/nar/28.1.316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Halfon MS, Gallo SM, Bergman CM. REDfly 2.0: an integrated database of cis-regulatory modules and transcription factor binding sites in Drosophila. Nucleic Acids Res. 2008;36:D594–D598. doi: 10.1093/nar/gkm876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Newburger DE, Bulyk ML. UniPROBE: an online database of protein binding microarray data on protein-DNA interactions. Nucleic Acids Res. 2009;37:D77–D82. doi: 10.1093/nar/gkn660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bailey TL, Boden M, Buske FA, Frith M, Grant CE, Clementi L, Ren J, Li WW, Noble WS. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Bailey TL, Williams N, Misleh C, Li WW. MEME: discovering and analyzing DNA and protein sequence motifs. Nucleic Acids Res. 2006;34:W369–W373. doi: 10.1093/nar/gkl198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Stormo GD. DNA binding sites: representation and discovery. Bioinformatics. 2000;16:16–23. doi: 10.1093/bioinformatics/16.1.16. [DOI] [PubMed] [Google Scholar]
- 32.Drysdale R. FlyBase : a database for the Drosophila research community. Methods Mol. Biol. 2008;420:45–59. doi: 10.1007/978-1-59745-583-1_3. [DOI] [PubMed] [Google Scholar]
- 33.Lyne R, Smith R, Rutherford K, Wakeling M, Varley A, Guillier F, Janssens H, Ji W, McLaren P, North P, et al. FlyMine: an integrated database for Drosophila and Anopheles genomics. Genome Biol. 2007;8:R129. doi: 10.1186/gb-2007-8-7-r129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Crooks GE, Hon G, Chandonia JM, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schneider TD, Stephens RM. Sequence logos: a new way to display consensus sequences. Nucleic Acids Res. 1990;18:6097–6100. doi: 10.1093/nar/18.20.6097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucl. Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Gupta S, Stamatoyannopoulos JA, Bailey TL, Noble WS. Quantifying similarity between motifs. Genome Biol. 2007;8:R24. doi: 10.1186/gb-2007-8-2-r24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Donlin MJ. Using the Generic Genome Browser (GBrowse) Curr. Protoc. Bioinformatics. 2009 doi: 10.1002/0471250953.bi0909s28. Chapter 9, Unit 9 9. [DOI] [PubMed] [Google Scholar]
- 39.Bailey TL, Elkan C. Fitting a mixture model by expectation maximization to discover motifs in biopolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol. 1994;2:28–36. [PubMed] [Google Scholar]
- 40.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bosch JRt, Benavides JA, Cline TW. The TAGteam DNA motif controls the timing of Drosophila pre-blastoderm transcription. Development. 2006;133:1967–1977. doi: 10.1242/dev.02373. [DOI] [PubMed] [Google Scholar]
- 42.Liang H-L, Nien C-Y, Liu H-Y, Metzstein MM, Kirov N, Rushlow C. The zinc-finger protein Zelda is a key activator of the early zygotic genome in Drosophila. Nature. 2008;456:400–403. doi: 10.1038/nature07388. [DOI] [PMC free article] [PubMed] [Google Scholar]