Abstract
We describe here OMPdb, which is currently the most complete and comprehensive collection of integral β-barrel outer membrane proteins from Gram-negative bacteria. The database currently contains 69 354 proteins, which are classified into 85 families, based mainly on structural and functional criteria. Although OMPdb follows the annotation scheme of Pfam, many of the families included in the database were not previously described or annotated in other publicly available databases. There are also cross-references to other databases, references to the literature and annotation for sequence features, like transmembrane segments and signal peptides. Furthermore, via the web interface, the user can not only browse the available data, but submit advanced text searches and run BLAST queries against the database protein sequences or domain searches against the collection of profile Hidden Markov Models that represent each family’s domain organization as well. The database is freely accessible for academic users at http://bioinformatics.biol.uoa.gr/OMPdb and we expect it to be useful for genome-wide analyses, comparative genomics as well as for providing training and test sets for predictive algorithms regarding transmembrane β-barrels.
INTRODUCTION
Integral membrane proteins account for ∼20–30% of fully sequenced proteomes (1). To date, two major structural architectures can be distinguished: the α-helical and the β-barrel membrane proteins. The former are located primarily in cell membranes of eukaryotic cells and bacterial inner membranes, while the latter are found exclusively in the outer membranes of Gram-negative bacteria and in the outer membranes of mitochondria and chloroplasts (2–4). The β-barrel outer membrane proteins (OMPs) are crucial for the life of bacteria, serving a variety of diverse roles, such as passive nutrient uptake and active transport of large molecules, protein secretion, enzymatic activity or adhesion to host cells (5–8).
The difficulty in obtaining crystals suitable for high-resolution studies of outer membrane proteins has resulted in their under-representation in the Protein Data Bank (9) (<1% of all deposited proteins with known 3D structure). Given these facts, and because many β-barrel OMPs nowadays attract an increased medical interest, several approaches have been made towards the development of predictive algorithms for this type of proteins. These methods are based grossly on hydrophobicity (10) and statistical analysis (11,12), remote homology detection (13), Hidden Markov Models (HMMs) (14–18), feed-forward Neural Networks (19–21), radial basis function Neural Networks (22,23) and Support Vector Machines (24), whereas others like BOMP (25), TMB-Hunt (26,27) and the TMB-finding pipeline (28) are oriented towards genome scale discrimination of β-barrel membrane proteins. A previously presented benchmark of several topology prediction methods indicated that HMMs are the most reliable predictors (29). Special purpose biological databases that include certain families of β-barrel proteins, like TCDB (30), PDBTM (31), TOPDB (32), PSORTdb (33), TMBETA-GENOME (34), OPM (35), Mptopo (36), Membrane Protein Data Bank (37), PRDNS (38), TMPDB (39), TMFunction (40) and the HHomp database, as part of the HHomp webserver (13) have also been available to the public. However, the annotation and classification of β-barrels in the aforementioned databases is incomplete, the coverage is limited and there are also many false positives (i.e. lipoproteins or peripheral proteins); this urges the need for intensive studies and careful data collection regarding these proteins.
MATERIALS AND METHODS
In order to create OMPdb, we based our research along three main axes, namely the available 3D structures, the profile HMMs (pHMMs) deposited in version 24.0 of the Pfam database (41) that correspond to domains found solely in transmembrane (TM) β-barrel OMPs, coupled with an extensive literature search for novel β-barrel proteins that are reported as such but cannot be retrieved from the databases otherwise. The ultimate purpose was to build a classification system where each family (i.e. a β-barrel domain) would be represented by a unique pHMM. We should emphasize here, that TM β-barrel proteins could possess either a single-domain architecture (where the whole sequence is composed of the β-barrel domain), or a multi-domain architecture (in which case the TM β-barrel domain comprises only a portion of the sequence). Thus, following the philosophy of domain databases such as Pfam, we identified only the respective domains that are responsible for the β-barrel formation and subsequently we included in the database only proteins possessing these domains.
Initially, we retrieved all proteins with a known 3D structure that are deposited in PDB and are listed at Stephen White’s laboratory page at UC Irvine (http://blanco.biomol.uci.edu/Membrane_Proteins_xtal.html). We chose to exclude β-barrels that do not originate from Gram-negative bacteria, such as the mitochondrial voltage-dependent anion channel (PDB ID: 2JK4) (42), the MspA mycobacterial outer membrane channel (PDB ID: 1UUN) (43) or the α-hemolysin (PDB ID: 7AHL) (44) and LukF (PDB ID:3LKF) (45) from Staphylococcus aureus. The retrieved proteins were subsequently matched against the pHMMs deposited in Pfam database. We studied the outer membrane β-barrel protein superfamily clan (MBB; CL0193) of the Pfam database which contains 36 members and noticed that there were several TM β-barrel proteins with crystallographically solved structure whose respective Pfam domains were not included in the clan. Such examples include ScrY (PDB ID:1A0S), OmpLA (PDB ID:1FW2) and PagP (PDB ID:1MM4), which correspond to Pfam domains PF02264, PF02253 and PF07017, respectively. Salmonella typhimurium LpxR (PDB ID:3FID) had a hit in a Pfam domain which was neither included in the clan nor had any description for its function (PF09982). Finally, two well-studied porins from Rhodobacter capsulatus (PDB ID:2POR) and Rhodopeudomonas blastica (PDB ID:1PRN) had no hits in the Pfam domains.
Apart from that, by searching in the literature, we came up with a number of proteins that were experimentally characterized as β-barrel (either multi- or single-domain) OMPs using several techniques (i.e. sub-cellular fractionization, electron microscopy, protease protection experiments, channel properties, chemical labeling, heat modifiable activity and so on). Many of them had either no hit in the pHMMs of Pfam or matched any of the automatically generated alignments in the Pfam-B subset. For these proteins there was also no evidence concerning their sub-cellular location or structure in the Uniprot database (46). In the first case, we performed a BLAST search (47) against Uniprot using as query the β-barrel domain of the experimentally verified protein(s) reported in the respective published reports, we created multiple alignments with the best-scoring results using ClustalW (48) and then, we built pHMMs for the particular domain using version 3.0b3 of the HMMER (49) software package. In the second case, we constructed the pHMMs based on the domain alignments deposited in Pfam-B after screening them in order to remove possible partial hits (sequence fragments).
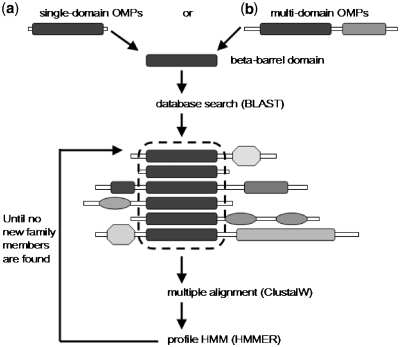
Finally, we used the total set of pHMMs that was created for retrieving additional proteins from Uniprot that scored high in these pHMMs, indicating that they could be classified in one of the available families. These proteins were appended in the multiple alignments representing the respective β-barrel domains and the procedure was repeated until no new members could be found. The procedure for creating the pHMM for the newly identified β-barrel domains is illustrated graphically in Figure 1. We have to notice, that some very short protein sequences (i.e. less than100 amino acids) that scored high against the pHMMs, were removed from the database since they could not possibly fold into a β-barrel structure.
Figure 1.
Schematic ilustration of the procedure used to create the pHMMs that represent the TM domains of β-barrel OMPs. Either we were dealing with (a) single-domain OMPs or (b) multi-domain ones, following the literature we isolated the domain responsible for the β-barrel formation. Afterwards, we performed a BLAST search (47) against Uniprot using as query the β-barrel domain of the experimentally verified protein reported in the respective published reports, we created multiple alignments with the best-scoring results using ClustalW (48) and then, we built pHMMs for the particular domain using version 3.0b3 of the HMMER (49) software package. The pHMM was subsequently used to search for new family members and the procedure iterated until no new family members are found. For families that were represented in Pfam-B subset, we used the already deposited automatically generated multiple alignments.
The majority of proteins that we collected would not be possible to be gathered by just searching in Uniprot using a combination of keywords, because they were mostly listed as having ‘putative’ or ‘unknown’ function. The result of the aforementioned procedure was a total of 85 families and 84 pHMMs (one family has only one representative, thus no multiple alignment and, consequently, no pHMM model could be created). At the end, we performed once again a literature search in order to find additional references for all the β-barrel families (i.e. domains) that we included in the database.
Another main innovation of OMPdb is the annotation of the TM segments. For proteins with known 3D structure, we used the information provided in the PDBTM database regarding the annotation of β-barrels along with the number and location of β-strands. For proteins that share structural homology with a protein with determined 3D structure (i.e. they belong to the same family), we mapped the boundaries of the TM segments of the member with the crystallographically solved structure to their sequences, using the alignment of the family’s sequences. Some obvious errors, such as a porin with 17 TM strands, were manually corrected. For families for which there was no evidence for TM topology, we offer the user the opportunity to run the PRED-TMBB algorithm, which is one of the top-scoring algorithms concerning β-barrel OMPs prediction (14,15,29).
RESULTS
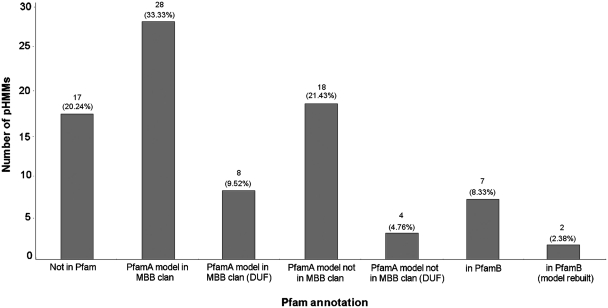
OMPdb, in its first version, contains 69 354 β-barrel outer membrane proteins, originating from 2712 Gram-negative bacterial species and strains, which are classified into 85 families. Figure 2 summarizes the annotation regarding the families’ classification system we propose in the Pfam database, which is the largest collection of protein families in the literature. However, if we just relied on the MBB clan, we would end up with only 36 profile HMM models that are identified by the curators of the database as being characteristic for β-barrels. A more detailed analysis, initiated by literature findings, revealed another 22 models deposited in Pfam-A, the high quality, manually curated component of Pfam, which represent β-barrels. Four out of them were actually assigned as domains of unknown function (DUFs), which lack further characterization in Pfam. We also used nine multiple sequence alignments (MSAs) from the automatically generated Pfam-B subset, because some of the proteins they contained were found to be β-barrels during the literature search. Both the 22 Pfam-A models and the nine MSAs from Pfam-B are not straightforward to be retrieved from Uniprot or Pfam just by using some kind of keywords combination in a text search query, since in most cases such keywords do not exist in the respective entries. More importantly, for 17 of OMPdb’s families, there was no information in the Pfam database at all. Figure 3 shows a summary of the database’s families and proteins entries classified according to their function.
Figure 2.
Annotation of OMPdb’s families in the latest version (v.24—October 2009) of the Pfam database. The majority of the families (36) is reported in the MBB clan part of the Pfam database. However, many pHMM models correspond to domains that represent families not included in the clan (18 + 4 = 22 families) or they can only be found in the non-annotated Pfam-B subset of Pfam (nine pHMMs). Finally, out of the 84 pHMM models that OMPdb contains, 17 were not reported in Pfam at all and these correspond to novel families.
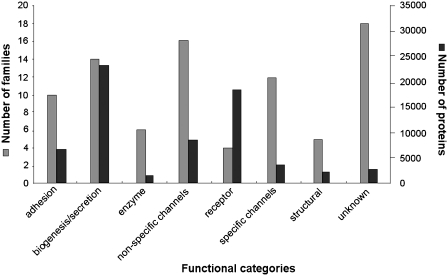
Figure 3.
A demonstration of the total numbers of families and protein entries in OMPdb that belong to each of the eight functional categories that we created. There are still a lot of families with β-barrel proteins whose function remains unclear, whereas specific and non-specific channels, together with biogenesis/secretion proteins seem to play a vital role in the bacterial life. Receptors are under-represented in terms of the number of families, but they constitute the second largest category of β-barrel OMPs in absolute numbers of proteins. In general, the largest proportion of outer membrane proteins in Gram-negative bacteria serve as receptors or play a role in biogenesis/secretion. A smaller amount comprises of non-specific channels and adhesion proteins. The category of proteins with unknown function, though it contains a lot of families, has a relatively small number of protein members, which may be due to the progress in whole genome annotation that is being made regarding Gram-negative bacteria.
The database possesses a user-friendly environment, through which the user may retrieve all the necessary information or find available resources and cross-references. The web application is based on the combination of two layers: the underlying level is a MySQL database system, which contains all protein data and the upper layer is an Apache-PHP applications server that receives user queries and fetches populated HTML data to the web browser client. From the welcome page of the web site, the user can view a brief description of OMPdb, along with the current holdings. A selection of a family entry from the drop down menu in the same page redirects to the respective family page. There, a short description of the family is presented along with the literature references, the initial (seed) and full alignments of the family’s members (Figure 4).
Figure 4.
Detailed view of a family entry. The user can read a short description for the given family together with a number of respective literature references. By clicking on the respective web links, he/she can view all proteins that belong to the family or download the seed and full alignments of the family’s protein members.
From the navigation bar in every web page, the user may choose one of the available search tools. There is a text search page, in which several fields are presented to the user, allowing the performance of advanced text search queries. This search can be limited in one of the available families as well. There is also the opportunity to submit BLAST searches against the database’s protein members using the BLAST search tool, where an E-value cut-off level can be specified accordingly. Additionally, through the Domain search tool, one or more FASTA-formatted sequences may be submitted and searched against the collection of profile HMMs that was constructed and is proven to be characteristic for β-barrel OMPs. This search can rely on either an E-value cut-off level or the manually specified trusted cut-off scores of each model. The results in all cases are presented in a tabular way, which facilitates easy view of the main fields of each result entry and can be selected and downloaded locally for further investigation. Each database entry contains the following fields: OMPdb name, OMPdb id, Uniprot accession number, protein description and classification, sequence, species, organism name, taxonomy, links to other databases, accompanied with annotation for TM segments and signal peptides (Figure 5). There is also an extensive user’s manual page, describing in detail the available tools.
Figure 5.
Detailed view of a protein entry of the database. The user may observe the classification of the protein, the available cross-references, the amino acid sequence, along with information about the presence of a signal peptide and the annotation of the TM segments. All this information can be downloaded in easy-to-use formats as well (FASTA, XML or TEXT files).
DISCUSSION
We have constructed a relational protein database, which contains β-barrel outer membrane proteins from Gram-negative bacteria. To our knowledge, OMPdb has some innovative and unique features not available in any other publicly accessible resource. When compared to general protein or domain databases, like Uniprot and Pfam (which were actually the main sources of information), OMPdb presents a more complete classification and accurate annotation concerning β-barrel proteins, due to the added value of the manual annotation that we performed and the detailed literature references that we provide.
On the other hand, a comparison of OMPdb against other specific databases that contain β-barrel proteins, reveals that it clearly excels at all aspects, because it features the largest number of protein and family entries, it possesses the most complete and exclusive data for β-barrels and offers the most complete interconnection to other public databases, literature references, prediction tools and sequence annotation. Table 1 shows a summary of the comparison of the main characteristics of OMPdb as opposed to other related databases that are available to the scientific community. From the table, it is evident that related databases fall into two main categories: there are those with a very limited number of protein entries and either no classification system for them or families with just few representative proteins. The reason for this is that they contain only proteins that have a crystallographically (3D) determined structured (like TCDB, PDB_TM, TOPDB, OPM, MPdb and TMPDB). On the other hand, there are databases like TMBETA-GENOME, PRNDS and PSORTdb that contain a larger number of proteins, but they offer no classification system and additionally, the proteins are gathered using automated techniques (i.e. prediction algorithms). We should note here, that prediction methods produce a large number of false positives and thus, the content of these databases need additional filtering (i.e. they contain several outer membrane lipoproteins or peripheral proteins). It is clear thus, that the semi-automated procedure used in the development of OMPdb, combines the advantages of both approaches offering currently the most advanced resource for OMPs.
Table 1.
A comparison of OMPdb to other databases that include β-barrel outer membrane proteins, in terms of total entries, method of data retrieval, existence of literature references, annotation of important sequence features (such as the signal peptide and the TM segments), interconnection to other public databases related to β-barrel proteins and availability of prediction/search tools
| Database features | OMPdb (current work) | PFAM (MBB clan) (41) | TCDB (30) | PDB_TM (nr set) (31) | TOPDB (32) | ePSORTdb (33) | TMBETA GENOME (34) | OPM (35) | MPdb (37) | PRNDS (38) | TMPDB (39) | HHompDB (13) |
|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Total entries | 69 354 | 38 595 | 348 | 49 | 45 | 480 | 36 095 | 73 | 148 | 3182 | 17 | 54 184 |
| Total families | 85 | 36 | 57 | − | − | − | − | 26 | 25 | − | − | − |
| Data retrieval | Semi- automated | Semi- automated | Manual | Semi- automated | Semi- automated | Manual | Automated | Semi- automated | Manual | Automated | Manual | Automated |
| Literature references | + | + | + | − | + | + | − | − | + | − | + | − |
| Signal peptide/ TM segments annotation | +/+ | −/− | −/− | −/+ | +/+ | −/− | −/− | −/+ | −/− | −/− | +/+ | −/− |
| Number of databases cross-referenced | 13 | 5 | 1 | 1 | 2 | 2 | − | 3 | 2 | 2 | 3 | − |
| Prediction/search tools | BLAST/ HMMER/ PRED-TMBB/ Text search | Text search/ HMMER | Text search/ BLAST | Text search | Text search | Psortb/ BLAST | Search by organism | Text search | Text search | Text search/ BSS/BBF | Text search | HHsearch |
OMPdb aims to fill a gap in the literature, because as already mentioned, the annotation of TM β-barrels in the existing databases is rather inadequate. Experimentalists that are involved with the biochemical and functional characterization of outer membrane proteins of Gram-negative bacteria will probably benefit the most from our database. For instance, a BLAST or Domain search query against OMPdb, which would follow the identification of one or more β-barrel protein sequences, is expected to be much more informative than a usual query against Uniprot, the non-redundant (nr) sequences of NCBI or Pfam. Moreover, even in the case that a similarity (even a remote one) is not found, the existing prediction methods that have been developed in our lab (PRED-TMBB and ConBBPRED), will provide the necessary tools towards the clarification of the structural and functional nature of these proteins.
Another aspect in which OMPdb could be very useful is the computational and theoretical analyses concerning β-barrels. The classification into families might be used in studies regarding the attributes of their members, in phylogenetic analyses and/or comparative genomics. The use of the pHMMs that are deposited in the database could also be used for designing effective strategies for whole-genome analyses.
Finally, the existence of such a large and reliable data set of β-barrels can be used for large-scale analyses concerning the classification accuracy of existing predictors, for training new prediction methods or for comparative modeling approaches. We permit the users to download the entire database, in various easy-to-use formats, for those bioinformaticians and/or experimentalists who would like to access a frequently updated, high-quality annotationed data set of β-barrel outer membrane proteins. We also used the CD-HIT program (50) in order to provide nr subsets of the database at various levels, i.e. 30% (4225 proteins), 50% (15 029 proteins), 70% (23 110 proteins) and 90% (31 327 proteins) sequence similarity. The database may also be useful for microbiologists whose aim is to identify potential targets in bacterial genomes for medical diagnostics, vaccines, antimicrobials and other uses.
Our long-term goal is to keep OMPdb as up-to-date as possible, following the regular updates of Uniprot and searching in the literature at the same time, for novel, experimentally verified β-barrel proteins, in order to include them in the database or appoint them to a new family if necessary. Similar to other databases, OMPdb is an ongoing project and interaction with the user community is vital for its development and refinement. We encourage the submission of data, correction of errors, and suggestions for making OMPdb of greater use to the scientific community.
FUNDING
Funding for the Open Access charge was provided by the authors.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank the anonymous referees whose comments helped in improving the quality of the article.
REFERENCES
- 1.Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 2.Cavalier-Smith T. Membrane heredity and early chloroplast evolution. Trends Plant Sci. 2000;5:174–182. doi: 10.1016/s1360-1385(00)01598-3. [DOI] [PubMed] [Google Scholar]
- 3.Elofsson A, von Heijne G. Membrane protein structure: prediction versus reality. Annu. Rev. Biochem. 2007;76:125–140. doi: 10.1146/annurev.biochem.76.052705.163539. [DOI] [PubMed] [Google Scholar]
- 4.Gray MW, Burger G, Lang BF. Mitochondrial evolution. Science. 1999;283:1476–1481. doi: 10.1126/science.283.5407.1476. [DOI] [PubMed] [Google Scholar]
- 5.Schulz GE. Transmembrane beta-barrel proteins. Adv. Protein Chem. 2003;63:47–70. doi: 10.1016/s0065-3233(03)63003-2. [DOI] [PubMed] [Google Scholar]
- 6.Wimley WC. The versatile beta-barrel membrane protein. Curr. Opin. Struct. Biol. 2003;13:404–411. doi: 10.1016/s0959-440x(03)00099-x. [DOI] [PubMed] [Google Scholar]
- 7.Schulz GE. Beta-barrel membrane proteins. Curr. Opin. Struct. Biol. 2000;10:443–447. doi: 10.1016/s0959-440x(00)00120-2. [DOI] [PubMed] [Google Scholar]
- 8.Gromiha MM, Suwa M. Current developments on beta-barrel membrane proteins: sequence and structure analysis, discrimination and prediction. Curr. Protein Pept. Sci. 2007;8:580–599. doi: 10.2174/138920307783018712. [DOI] [PubMed] [Google Scholar]
- 9.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Zhai Y, Saier MH., Jr The beta-barrel finder (BBF) program, allowing identification of outer membrane beta-barrel proteins encoded within prokaryotic genomes. Protein Sci. 2002;11:2196–2207. doi: 10.1110/ps.0209002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Freeman TC, Jr, Wimley WC. A highly accurate statistical approach for the prediction of transmembrane beta-barrels. Bioinformatics. 2010;26:1965–1974. doi: 10.1093/bioinformatics/btq308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wimley WC. Toward genomic identification of beta-barrel membrane proteins: composition and architecture of known structures. Protein Sci. 2002;11:301–312. doi: 10.1110/ps.29402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Remmert M, Linke D, Lupas AN, Soding J. HHomp–prediction and classification of outer membrane proteins. Nucleic Acids Res. 2009;37:W446–W451. doi: 10.1093/nar/gkp325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. A Hidden Markov Model method, capable of predicting and discriminating beta-barrel outer membrane proteins. BMC Bioinformatics. 2004;5:29. doi: 10.1186/1471-2105-5-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bagos PG, Liakopoulos TD, Spyropoulos IC, Hamodrakas SJ. PRED-TMBB: a web server for predicting the topology of beta-barrel outer membrane proteins. Nucleic Acids Res. 2004;32:W400–W404. doi: 10.1093/nar/gkh417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bigelow HR, Petrey DS, Liu J, Przybylski D, Rost B. Predicting transmembrane beta-barrels in proteomes. Nucleic Acids Res. 2004;32:2566–2577. doi: 10.1093/nar/gkh580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liu Q, Zhu YS, Wang BH, Li YX. A HMM-based method to predict the transmembrane regions of beta-barrel membrane proteins. Comput. Biol. Chem. 2003;27:69–76. doi: 10.1016/s0097-8485(02)00051-7. [DOI] [PubMed] [Google Scholar]
- 18.Martelli PL, Fariselli P, Krogh A, Casadio R. A sequence-profile-based HMM for predicting and discriminating beta barrel membrane proteins. Bioinformatics. 2002;18(Suppl. 1):S46–S53. doi: 10.1093/bioinformatics/18.suppl_1.s46. [DOI] [PubMed] [Google Scholar]
- 19.Diederichs K, Freigang J, Umhau S, Zeth K, Breed J. Prediction by a neural network of outer membrane beta-strand protein topology. Protein Sci. 1998;7:2413–2420. doi: 10.1002/pro.5560071119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Gromiha MM, Ahmad S, Suwa M. Neural network-based prediction of transmembrane beta-strand segments in outer membrane proteins. J. Comput. Chem. 2004;25:762–767. doi: 10.1002/jcc.10386. [DOI] [PubMed] [Google Scholar]
- 21.Jacoboni I, Martelli PL, Fariselli P, De Pinto V, Casadio R. Prediction of the transmembrane regions of beta-barrel membrane proteins with a neural network-based predictor. Protein Sci. 2001;10:779–787. doi: 10.1110/ps.37201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ou YY, Chen SA, Gromiha MM. Prediction of membrane spanning segments and topology in beta-barrel membrane proteins at better accuracy. J. Comput. Chem. 2010;31:217–223. doi: 10.1002/jcc.21281. [DOI] [PubMed] [Google Scholar]
- 23.Ou YY, Gromiha MM, Chen SA, Suwa M. TMBETADISC-RBF: discrimination of beta-barrel membrane proteins using RBF networks and PSSM profiles. Comput. Biol. Chem. 2008;32:227–231. doi: 10.1016/j.compbiolchem.2008.03.002. [DOI] [PubMed] [Google Scholar]
- 24.Natt NK, Kaur H, Raghava GP. Prediction of transmembrane regions of beta-barrel proteins using ANN- and SVM-based methods. Proteins. 2004;56:11–18. doi: 10.1002/prot.20092. [DOI] [PubMed] [Google Scholar]
- 25.Berven FS, Flikka K, Jensen HB, Eidhammer I. BOMP: a program to predict integral beta-barrel outer membrane proteins encoded within genomes of Gram-negative bacteria. Nucleic Acids Res. 2004;32:W394–W399. doi: 10.1093/nar/gkh351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Garrow AG, Agnew A, Westhead DR. TMB-Hunt: a web server to screen sequence sets for transmembrane beta-barrel proteins. Nucleic Acids Res. 2005;33:W188–W192. doi: 10.1093/nar/gki384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Garrow AG, Westhead DR. A consensus algorithm to screen genomes for novel families of transmembrane beta barrel proteins. Proteins. 2007;69:8–18. doi: 10.1002/prot.21439. [DOI] [PubMed] [Google Scholar]
- 28.Gromiha MM, Yabuki Y, Suwa M. TMB finding pipeline: novel approach for detecting beta-barrel membrane proteins in genomic sequences. J. Chem. Inf. Model. 2007;47:2456–2461. doi: 10.1021/ci700222s. [DOI] [PubMed] [Google Scholar]
- 29.Bagos PG, Liakopoulos TD, Hamodrakas SJ. Evaluation of methods for predicting the topology of beta-barrel outer membrane proteins and a consensus prediction method. BMC Bioinformatics. 2005;6:7. doi: 10.1186/1471-2105-6-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Saier MH, Jr, Tran CV, Barabote RD. TCDB: the Transporter Classification Database for membrane transport protein analyses and information. Nucleic Acids Res. 2006;34:D181–D186. doi: 10.1093/nar/gkj001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tusnady GE, Dosztanyi Z, Simon I. PDB_TM: selection and membrane localization of transmembrane proteins in the protein data bank. Nucleic Acids Res. 2005;33:D275–D278. doi: 10.1093/nar/gki002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tusnady GE, Kalmar L, Simon I. TOPDB: topology data bank of transmembrane proteins. Nucleic Acids Res. 2008;36:D234–D239. doi: 10.1093/nar/gkm751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Rey S, Acab M, Gardy JL, Laird MR, deFays K, Lambert C, Brinkman FS. PSORTdb: a protein subcellular localization database for bacteria. Nucleic Acids Res. 2005;33:D164–D168. doi: 10.1093/nar/gki027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gromiha MM, Yabuki Y, Kundu S, Suharnan S, Suwa M. TMBETA-GENOME: database for annotated beta-barrel membrane proteins in genomic sequences. Nucleic Acids Res. 2007;35:D314–D316. doi: 10.1093/nar/gkl805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Lomize MA, Lomize AL, Pogozheva ID, Mosberg HI. OPM: orientations of proteins in membranes database. Bioinformatics. 2006;22:623–625. doi: 10.1093/bioinformatics/btk023. [DOI] [PubMed] [Google Scholar]
- 36.Jayasinghe S, Hristova K, White SH. MPtopo: a database of membrane protein topology. Protein Sci. 2001;10:455–458. doi: 10.1110/ps.43501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Raman P, Cherezov V, Caffrey M. The Membrane Protein Data Bank. Cell Mol. Life Sci. 2006;63:36–51. doi: 10.1007/s00018-005-5350-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Katta AV, Marikkannu R, Basaiawmoit RV, Krishnaswamy S. Consensus based validation of membrane porins. In Silico Biol. 2004;4:549–561. [PubMed] [Google Scholar]
- 39.Ikeda M, Arai M, Okuno T, Shimizu T. TMPDB: a database of experimentally-characterized transmembrane topologies. Nucleic Acids Res. 2003;31:406–409. doi: 10.1093/nar/gkg020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Gromiha MM, Yabuki Y, Suresh MX, Thangakani AM, Suwa M, Fukui K. TMFunction: database for functional residues in membrane proteins. Nucleic Acids Res. 2009;37:D201–D204. doi: 10.1093/nar/gkn672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bayrhuber M, Meins T, Habeck M, Becker S, Giller K, Villinger S, Vonrhein C, Griesinger C, Zweckstetter M, Zeth K. Structure of the human voltage-dependent anion channel. Proc. Natl Acad. Sci. USA. 2008;105:15370–15375. doi: 10.1073/pnas.0808115105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Faller M, Niederweis M, Schulz GE. The structure of a mycobacterial outer-membrane channel. Science. 2004;303:1189–1192. doi: 10.1126/science.1094114. [DOI] [PubMed] [Google Scholar]
- 44.Song L, Hobaugh MR, Shustak C, Cheley S, Bayley H, Gouaux JE. Structure of staphylococcal alpha-hemolysin, a heptameric transmembrane pore. Science. 1996;274:1859–1866. doi: 10.1126/science.274.5294.1859. [DOI] [PubMed] [Google Scholar]
- 45.Olson R, Nariya H, Yokota K, Kamio Y, Gouaux E. Crystal structure of staphylococcal LukF delineates conformational changes accompanying formation of a transmembrane channel. Nat. Struct. Biol. 1999;6:134–140. doi: 10.1038/5821. [DOI] [PubMed] [Google Scholar]
- 46.Bairoch A, Apweiler R, Wu CH, Barker WC, Boeckmann B, Ferro S, Gasteiger E, Huang H, Lopez R, Magrane M, et al. The Universal Protein Resource (UniProt) Nucleic Acids Res. 2005;33:D154–D159. doi: 10.1093/nar/gki070. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Thompson JD, Gibson TJ, Higgins DG. Multiple sequence alignment using ClustalW and ClustalX. Curr. Protoc. Bioinformatics. 2002 doi: 10.1002/0471250953.bi0203s00. Chapter 2, Unit 2. 3. [DOI] [PubMed] [Google Scholar]
- 49.Eddy SR. Profile hidden Markov models. Bioinformatics. 1998;14:755–763. doi: 10.1093/bioinformatics/14.9.755. [DOI] [PubMed] [Google Scholar]
- 50.Li W, Godzik A. Cd-hit: a fast program for clustering and comparing large sets of protein or nucleotide sequences. Bioinformatics. 2006;22:1658–1659. doi: 10.1093/bioinformatics/btl158. [DOI] [PubMed] [Google Scholar]