Abstract
The International Knockout Mouse Consortium (IKMC) aims to mutate all protein-coding genes in the mouse using a combination of gene targeting and gene trapping in mouse embryonic stem (ES) cells and to make the generated resources readily available to the research community. The IKMC database and web portal (www.knockoutmouse.org) serves as the central public web site for IKMC data and facilitates the coordination and prioritization of work within the consortium. Researchers can access up-to-date information on IKMC knockout vectors, ES cells and mice for specific genes, and follow links to the respective repositories from which corresponding IKMC products can be ordered. Researchers can also use the web site to nominate genes for targeting, or to indicate that targeting of a gene should receive high priority. The IKMC database provides data to, and features extensive interconnections with, other community databases.
INTRODUCTION
The mouse serves as a premier animal model in biomedical research because it is closely related to human and because it is readily accessible to detailed genetic, molecular and phenotypic analysis. With the availability of the mouse genome sequence, proposals were put forward to systematically mutate all mouse protein-coding genes in ES cells and thus to create, in a highly time and cost-effective manner, a valuable resource of genetic mutations for future biomedical research (1,2). In response, three major mouse knockout programs were initiated: KOMP (KnockOut Mouse Project) funded by the National Institutes of Health (NIH, USA); EUCOMM (EUropean Conditional Mouse Mutagenesis Program) funded by the European Commission (EC) and NorCOMM (North American Conditional Mouse Mutagenesis Project) funded by Genome Canada. These three programs formed the International Knockout Mouse Consortium (3), and a fourth group, the Texas Institute of Genomic Medicine (TIGM), soon joined this large-scale collaborative effort (4).
The IKMC web portal (www.knockoutmouse.org) was developed and is being maintained jointly by the KOMP-DCC (KOMP-Data Coordination Center) project funded by the National Institutes of Health, and the I-DCC (International-Data Coordination Center) project funded by the EC. It is the official and central entry point to all IKMC data and resources. The primary objectives of the IKMC web portal and database are to (i) provide infrastructure to select and prioritize genes to be targeted across IKMC production centers, (ii) facilitate coordination of work and to track progress within the IKMC and (iii) make all the data readily accessible to the research community, together with links to the repositories which distribute IKMC products. KOMP targeting vectors, ES cells and mice are being distributed by the KOMP Repository (www.komp.org); EUCOMM vectors and ES cell lines by the European Mouse Mutant Cell Repository (www.eummcr.org); EUCOMM mice by the European Mouse Mutant Archive (EMMA) (5); NorCOMM targeting vectors, ES cells and mice by the Toronto Centre for Phenogenomics (www.phenogenomics.ca) and TIGM gene trap ES cells and mice from TIGM (6).
THE IKMC DATABASE
The unified and annotated master gene list
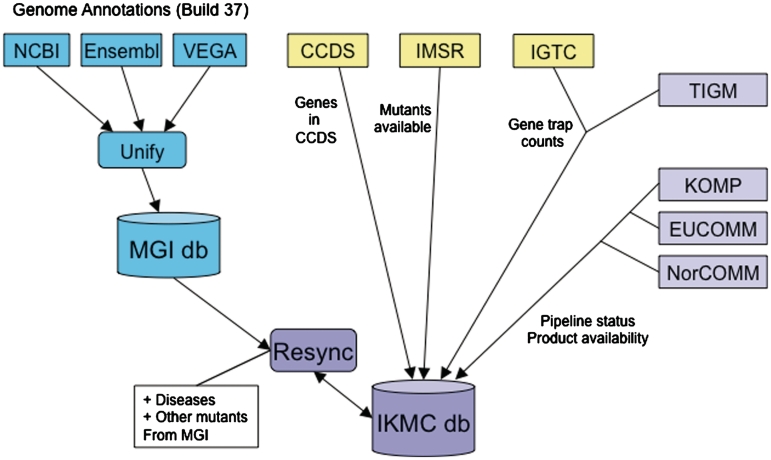
The design of gene targeting vectors requires detailed knowledge about gene models, i.e. about the intron/exon structure of genes. While there are many cases in which there is a one-to-one mapping of gene predictions from different genome annotation pipelines (Ensembl, Vega, NCBI) (7–9), there are also a substantial number of genes for which the gene models don't agree. There are genes that are only predicted by one resource, or genomic regions for which one pipeline predicts one and another several gene models. Therefore, the comparison, coordination and integration of IKMC efforts require one unified catalog of mouse genes and gene models (Figure 1). A genome feature catalog for the reference mouse genome that combines the genome annotations from Ensembl, Havana and NCBI is maintained at MGI (10). The resolution of conflicting annotation is accomplished through close and ongoing collaboration between members of the MGI, Havana, Ensembl and NCBI genome annotation and curation teams. The IKMC database takes advantage of and contributes to this work. The IKMC gene list is synchronized with the MGI gene catalog daily, thus providing the basis for coordination and integrated querying capabilities. The genes on the unified master list are then annotated with information from various external resources that helps to coordinate and prioritize work within and between IKMC projects. Annotations include, for example, genes belonging to the CCDS (Consensus Coding Sequence Set) (11) and thus having high-confidence gene models, genes with human orthologs with disease entries in OMIM (12), genes trapped by the IGTC (International Gene Trap Consortium) (13) or by TIGM, genes for which mutants are reported in MGI, and genes for which mutant ES cells or mice are available through the International Mouse Strain Resource (IMSR) (14).
Figure 1.
Schematic of information flow into the IKMC database. The IKMC database and web portal facilitate gene selection, prioritization and coordination among IKMC participants (purple shading). Data are updated daily. The master gene list for IKMC is maintained by synchronizing with genes represented in the MGI database. The MGI non-redundant mouse gene catalog is built by comparing and unifying genome annotations from Ensembl, Vega and NCBI (blue shading). Gene records in the IKMC database are then annotated with additional information from many resources, including MGI, NCBI (CCDS), the IMSR, the IGTC and the IKMC production centers and repositories.
Based on all the annotation criteria, computational utilities are provided to the IKMC project leaders that facilitate coordinated gene selection and assignment of targeting projects to specific KOMP, EUCOMM and NorCOMM production centers. The IKMC database tracks gene assignments, targeting projects and their statuses for each gene in every production pipeline, and product availability from IKMC repositories. All this information, as well as pertinent links to external resources, is updated daily and readily available via the public IKMC web interface (see below).
Targeting vector and mutant allele information
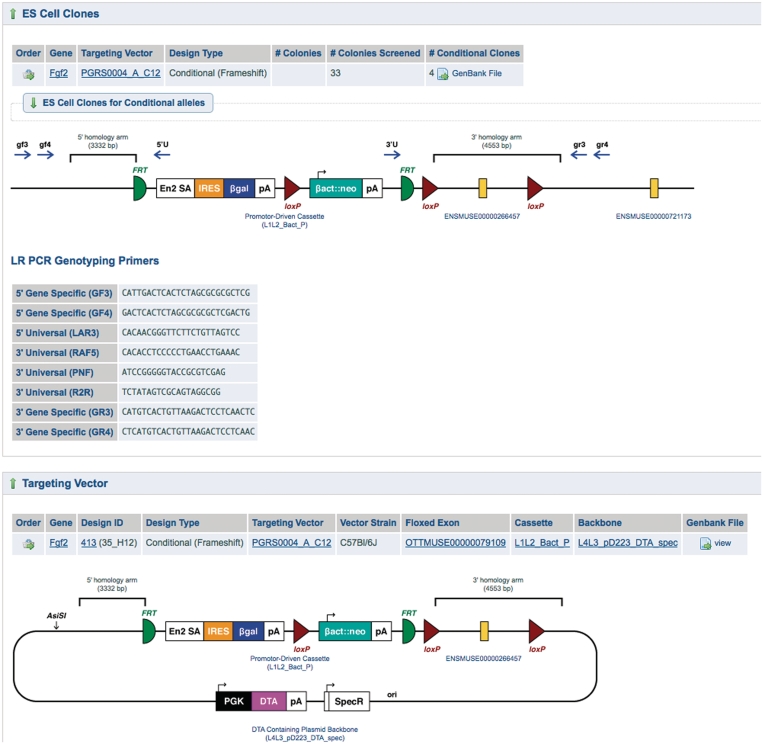
The IKMC Targeting repository is a component of the IKMC database, which stores a catalog of available products and nucleotide-level descriptions of mutant alleles generated by the IKMC program. Program participants submit data directly on the internal website visible at www.knockoutmouse.org/targ_rep, or programmatically upload data in bulk into the repository. For every targeting vector and ES cell clone entered into the catalog, the repository stores and presents annotated mutant allele sequence, a downloadable image of the targeting vector and mutant allele, and any QC performed on mutant ES cells by the distribution centers and mouse production facilities. The mutant allele sequence is intended to help researchers design their own experiments and verify the IKMC products they receive. The images are of a quality that can be used in publications, and the QC information stored for ES cell clones make it clear which assays have been performed on the clones, and thus, the level of due diligence expected of the end-user.
This mutant allele information is stored and presented in the ‘same’ uniform format for all IKMC mutants, regardless of knockout program. The information is made publicly available in three places: the ‘details’ section of the IKMC web portal (see below), the allele pages (Figure 4), and a DAS-Track for any client, including the Ensembl genome browser.
Figure 4.
Screenshot showing the detailed targeting vector and allele information provided at the IKMC web site. The example shown is a conditional-ready/knockout-first allele (3,20). The salient molecular features of the targeting vector and the resulting mutant allele in ES cells are displayed, including homology arms, the FRT and loxP sites and the location of primers used for quality controls. Ensembl exon IDs link to genome coordinate information. Links to GenBank files provide targeting vector and mutant allele information at the sequence level.
THE IKMC WEB PORTAL: ACCESSING IKMC DATA AND RESOURCES
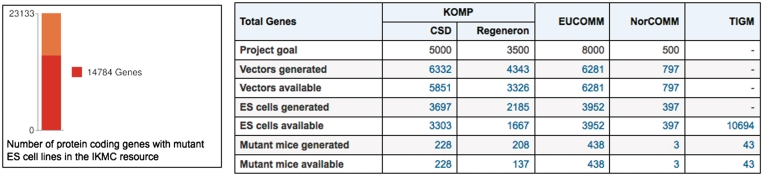
The IKMC web site (www.knockoutmouse.org) is a central entry point to IKMC data and resources. The portal presents general information about each consortium member and about IKMC allele types and targeting strategies. The home page provides a table summarizing current progress and downloadable data reports. The status of the IKMC initiative, as of August 2010, is shown in Figure 2. Most importantly, the web site features search and browse functions that allow researchers to determine whether their genes of interest are being targeted by the IKMC and the current status of these projects. Users can readily look up the detailed molecular structure of corresponding targeting vectors and mutant alleles, and which targeting vectors, ES cells or mice are available to order.
Figure 2.
Summary of the progress for the IKMC effort, as of August 2010. To date, mutant ES cell lines for 14 737 protein-coding genes have been generated by the IKMC. The table shows the number of genes for which targeting vectors, mutant ES cells and mutant mice are ready for distribution.
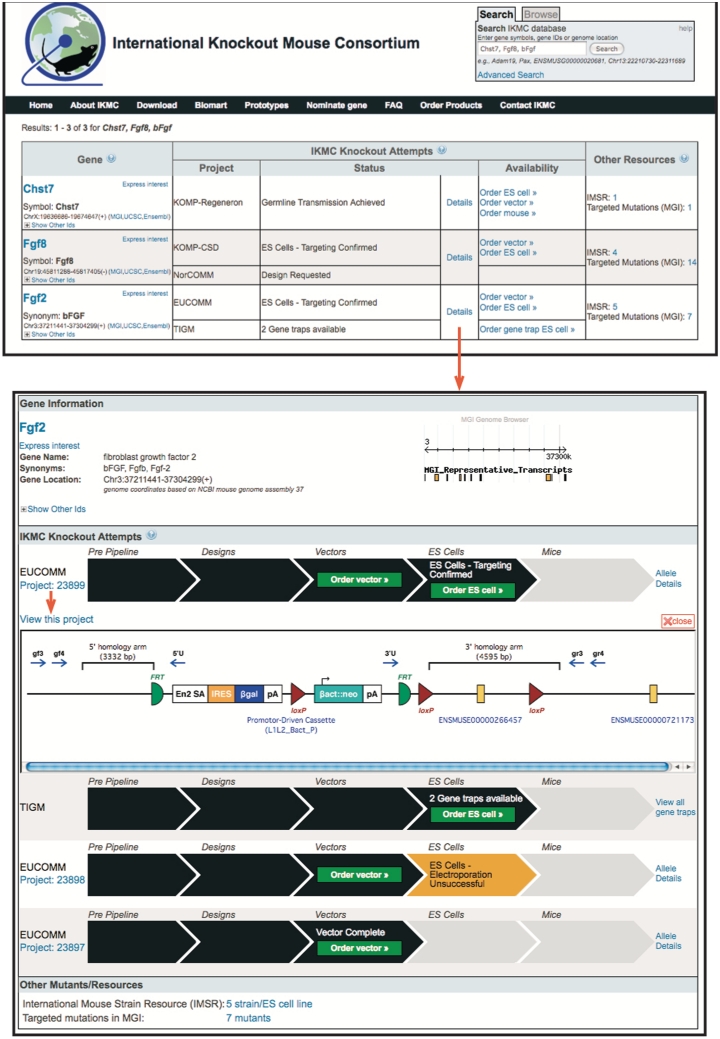
Queries return tabular summaries that list one record for each gene that matches the query (Figure 3). Each record includes a high-level summary of IKMC knockout attempts, indicating which programs are working on the gene and the status of the most advanced targeting effort per program with links to more details. The availability of targeting vectors, mutant ES cells and mice and links to the repositories that distribute the respective IKMC products are also provided. In addition, query summaries list other resources (IMSR, IGTC and MGI) that report on additional mutant ES cells or mice for the gene of interest, with the number of, and links to, corresponding entries at the respective sites.
Figure 3.
Query summary and detail pages. Query summaries (top) display one record for each gene matching the query. One can query by gene symbols, by various gene IDs and by genome coordinates. Matches to synonyms are returned as well and the reason for matching is indicated in the gene column. The gene column displays the official gene symbol, linked to the corresponding gene pages at MGI. Gene specific links to genome browsers and other external sites are provided. By following the ‘Express interest’ link, researchers can provide input on gene selection and prioritization for targeting experiments. The IKMC Knockout Attempts column lists all IKMC programs working on the gene. The status of the most advanced targeting effort per program is shown, with a link to more details. The availability of IKMC products is indicated by order links to the respective IKMC repositories. The Other Resources column displays the number of mutant ES cells and mice reported by the IMSR, IGTC and MGI with links to the corresponding entries at these sites. The Details page (bottom) lists all the IKMC knockout attempts for a given gene. The most advanced projects are displayed first. Available products are indicated by order links to the respective repositories. Clicking on the ‘Allele Details’ link (to the right of the project status bar) opens a graphical display illustrating the features of the mutant allele generated by the project. The ‘View this project’ link leads to more comprehensive targeting vector and mutant allele information (Figure 4).
The ‘Details’ page (Figure 3) lists all IKMC knockout attempts for the gene of interest. Different targeting strategies may be employed to mutate a gene, resulting in several targeting projects. The production pipeline status for each project is displayed, and the availability of products is indicated via order links to the respective repositories. As soon as a targeting vector has been built for a specific project, links to detailed vector and mutant allele information are provided as well. These allele pages (currently available for KOMP and EUCOMM; NorCOMM will be added soon) illustrate the molecular features of targeting vectors and describe the mutations of ES cells at the sequence level (Figure 4). Thus, researchers can determine the exact nature of targeting vectors and mutant ES cells and mice before they proceed to ordering specific IKMC products.
COMMUNITY INPUT ON GENE SELECTION
Researchers are encouraged to nominate genes for targeting, or to request a high priority for a particular gene. This can be done by using the ‘Nominate gene’ utility on the navigation bar of each IKMC web portal page, and by clicking the ‘Nominate’ or ‘Express interest’ link displayed in individual gene records. As of 4 August 2010, 1377 nominations for 1188 genes have been received from the scientific community.
EXPORT OF IKMC DATA TO OTHER COMMUNITY RESOURCES
The IKMC database provides regularly updated GFF files of mutant allele information to the UCSC- (15) and MGI genome browsers for the display of IKMC allele tracks. In addition, the targeting repository serves a DAS-track (via the Wellcome Trust Sanger Institute DAS-server) to any interested client, in particular the Ensembl web browser. The allele tracks on the UCSC, MGI and ENSEMBL genome browsers are color-coded to indicate the status for each targeting project and link back to more information in the IKMC web portal. IKMC allele information is registered in MGI and official allele IDs and nomenclature are established in collaboration with MGI. Official allele IDs provide important integration points for biological data that will come from the studies of IKMC targeted ES cells and mice. Downloadable reports of all KOMP and EUCOMM targeted alleles are available from the IKMC web portal (http://www.knockoutmouse.org/download) and from MGI's ftp site (ftp://ftp.informatics.jax.org/pub/reports/index.html#pheno).
USER SUPPORT
The IKMC web portal supports its users through on-line documentation and a dedicated User Support staff. The on-line documentation is accessible via the FAQ link in the navigation bar and by clicking on question marks that are displayed on web pages. Our User Support personnel can be reached by using the ‘Contact IKMC’ utility in the navigation bar.
FUTURE DEVELOPMENTS
The IKMC database and web site will continue to evolve. We will continue to expand and enhance the representation of vector and mutant allele information. Currently, this information is available for targeted mutants from the EUCOMM and KOMP programs, in a format that combines graphical views and sequence feature files. Including targeted allele information from NorCOMM, and adding a graphical representation of mutant alleles in their genomic context are important pending tasks. We are also completing the storage of targeted allele information from other large-scale targeting programs such as the Sanger Institute microRNA Knockout program (MirKO) and epitope-tagged alleles generated by the EUTRACC consortium (www.eutracc.eu). Our intent is to expand the targeting repository to include all future, publically-available targeted alleles in the mouse generated from large-scale programs.
The characterization of IKMC gene trap alleles by the TIGM, EUCOMM and NorCOMM programs, as well as other gene traps with sequence tags deposited in the dbGSS library, is now nearing completion, in the form of a Gene Trap Data Repository. Gene trap alleles are characterized using a modified Unitrap (16) algorithm for sequence-tag mapping and clustering. The definition of these gene-trap alleles not only simplifies the presentation of gene-trap data by condensing multiple gene traps into clusters that express the same fusion transcript, but also allows us to provide mutant allele sequence and graphics using the same methods already developed for targeted mutations. When complete, the Gene Trap Data Repository will form another component of the IKMC database, and its allele information will be served by the IKMC portal alongside the information on targeted mutations.
One particularly exciting aspect of our current and future work is to integrate IKMC targeted alleles with additional biological information from other sources through the use of BioMart technology (17). A BioMart portal prototype is available on the ‘Prototypes’ tab of the IKMC web site (www.knockoutmouse.org/martsearch). Currently, this portal combines information on IKMC mouse knockout resources with numerous other relevant datasets, including gene information from MGI and Ensembl, gene expression data from EurExpress (www.eurexpress.org), phenotype data from Europhenome (18), and mouse distribution information from EMMA. Work is underway to develop BioMarts of GXD gene expression information (19), biochemical pathway and human disease associations. Data representation, integration and querying capabilities via the BioMart interface will be refined and data from other resources will be added. This feature will further enhance the utility of IKMC data and help to realize the enormous potential of IKMC resources for future biomedical research.
TECHNICAL INFORMATION
The IKMC database is implemented in PostgreSQL version 8.3.7. The software to resynchronize the IKMC gene list with the MGI gene catalog, load annotations and load pipeline statuses is written in Python and Java using Hibernate. The web site static content is served using Drupal, an open source content management platform, and the dynamic search features are implemented using PHP and an Apache Solr/Lucene index. The index is refreshed from the PostgreSQL database daily.
The Targeting Repository (http://www.knockoutmouse/targ_rep) is written in Ruby using the Ruby on Rails application framework, with a MySQL database back end. The prototype BioMart portal (at http://www.knockoutmouse.org/martsearch) is written in Ruby using the Sinatra application framework and the search-engine component is powered by Apache Solr as well as the individual BioMarts.
FUNDING
European Commission: Project number 223592; National Institutes of Health, National Human Genome Research Institute: Grant number HG004074. Funding for open access charge: European Commission: Project number 223592; National Institutes of Health grant HG004074.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank all our colleagues from the different IKMC programs for making their data available to the IKMC web portal project and for very collegial and productive interactions. Specific thanks go to Infejinelo Onyiah for his contributions to the code that draws the vector and allele images. We would like to thank the members of the IKMC steering committee and the advisors of the IKMC programs for providing valuable input and feedback on the development of the IKMC web portal. Finally, we would like to thank UCSC and Ensembl for displaying IKMC allele tracks on their genome browsers, and all external resources that provide data to the IKMC web portal.
REFERENCES
- 1.Austin CP, Battey JF, Bradley A, Bucan M, Capecchi M, Collins FS, Dove WF, Duyk G, Dymecki S, Eppig JT, et al. The knockout mouse project. Nature Genet. 2004;36:921–924. doi: 10.1038/ng0904-921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Auwerx J, Avner P, Baldock R, Ballabio A, Balling R, Barbacid M, Berns A, Bradley A, Brown S, Carmeliet P, et al. The European dimension for the mouse genome mutagenesis program. Nature Genet. 2004;36:925–927. doi: 10.1038/ng0904-925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.International Mouse Knockout Consortium; Collins FS, Rossant J, Wurst W. A mouse for all reasons. Cell. 2007;128:9–13. doi: 10.1016/j.cell.2006.12.018. [DOI] [PubMed] [Google Scholar]
- 4.Collins FS, Finnell RH, Rossant J, Wurst W. A new partner for the international knockout mouse consortium. Cell. 2007;129:235. doi: 10.1016/j.cell.2007.04.007. [DOI] [PubMed] [Google Scholar]
- 5.Wilkinson P, Sengerova J, Matteoni R, Chen CK, Soulat G, Ureta-Vidal A, Fessele S, Hagn M, Massimi M, Pickford K, et al. EMMA – mouse mutant resources for the international scientific community. Nucleic Acids Res. 2010;38:D570–D576. doi: 10.1093/nar/gkp799. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hansen GM, Markesich DC, Burnett MB, Zhu Q, Dionne KM, Richter LJ, Finnell RH, Sands AT, Zambrowicz BP, Abuin A. Large-scale gene trapping in C57BL/6N mouse embryonic stem cells. Genome Res. 2008;18:1670–1679. doi: 10.1101/gr.078352.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT Mouse Genome Database Group. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Nord AS, Chang PJ, Conklin BR, Cox AV, Harper CA, Hicks GG, Huang CC, Johns SJ, Kawamoto M, Liu S, et al. The International Gene Trap Consortium Website: a portal to all publicly available gene trap cell lines in mouse. Nucleic Acids Res. 2006;34:D642–D648. doi: 10.1093/nar/gkj097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Eppig JT, Strivens M. Finding a mouse: the International Mouse Strain Resource (IMSR) Trends Genet. 1999;15:81–82. doi: 10.1016/s0168-9525(98)01665-5. [DOI] [PubMed] [Google Scholar]
- 15.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roma G, Sardiello M, Cobellis G, Cruz P, Lago G, Sanges R, Stupka E. The UniTrap resource: tools for the biologist enabling optimized use of gene trap clones. Nucleic Acids Res. 2008;36:D741–746. doi: 10.1093/nar/gkm825. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart – biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Morgan H, Beck T, Blake A, Gates H, Adams N, Debouzy G, Leblanc S, Lengger C, Maier H, Melvin D, et al. EuroPhenome: a repository for high-throughput mouse phenotyping data. Nucleic Acids Res. 2010;38:D577–585. doi: 10.1093/nar/gkp1007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Smith CM, Finger JH, Hayamizu TF, McCright IJ, Eppig JT, Kadin JA, Richardson JE, Ringwald M. The mouse Gene Expression Database (GXD): 2007 update. Nucleic Acids Res. 2007;35:D618–623. doi: 10.1093/nar/gkl1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Testa G, Schaft J, van der Hoeven F, Glaser S, Anastassiadis K, Zhang Y, Hermann T, Stremmel W, Stewart AF. A reliable lacZ expression reporter cassette for multipurpose, knockout-first alleles. Genesis. 2004;38:151–158. doi: 10.1002/gene.20012. [DOI] [PubMed] [Google Scholar]