Abstract
Cryo-electron microscopy reconstruction methods are uniquely able to reveal structures of many important macromolecules and macromolecular complexes. EMDataBank.org, a joint effort of the Protein Data Bank in Europe (PDBe), the Research Collaboratory for Structural Bioinformatics (RCSB) and the National Center for Macromolecular Imaging (NCMI), is a global ‘one-stop shop’ resource for deposition and retrieval of cryoEM maps, models and associated metadata. The resource unifies public access to the two major archives containing EM-based structural data: EM Data Bank (EMDB) and Protein Data Bank (PDB), and facilitates use of EM structural data of macromolecules and macromolecular complexes by the wider scientific community.
INTRODUCTION
Cryo-electron microscopy (cryoEM) has become an essential technique in structural biology, bridging the gap between cell biology, X-ray crystallography and nuclear magnetic resonance (NMR) spectroscopy (1,2). CryoEM reconstruction methods are being used to determine structures of large macromolecules, macromolecular complexes and cell components involved in many key biological processes including signal transduction, genome replication, transcription, translation, chaperonin-assisted protein folding, viral infection and motility. Three-dimensional (3D) density maps derived from cryoEM experiments reveal overall molecular shape and may be further interpreted through segmentation algorithms, rigid-body fitting of atomic coordinates determined using X-ray crystallography or NMR, and/or ab initio model building, depending on map resolution (3–5).
Public access to cryoEM map volumes and their fitted model interpretations permits independent assessment and analysis of structural results and stimulates development of new tools for visualization, fitting and validation. The EM Data Bank (EMDB) is the major repository for 3D map volumes obtained using electron microscopy (6), while the Protein Data Bank (PDB) collects atomic coordinates fitted into EM map volumes (7). The Unified Data Resource for CryoEM (http://EMDataBank.org) was created in order to unify data deposition, processing and retrieval of maps and fitted models. Here, we provide an overview of the EM structural data archives and the unified resource, including their historical context, current content and use, and future prospects.
EM STRUCTURAL DATA ARCHIVES
Maps
The EMDB was established in 2002 by the Macromolecular Structure Database group (now PDBe) at the European Bioinformatics Institute (EBI) in Hinxton, UK (6,8,9), and was initially supported by two European Union-funded projects: Integration of Information about Macromolecular Structure (IIMS) and the 3D Electron Microscopy Network of Excellence (3DEM NoE). A web-based deposition system, EMDEP, was developed to handle data capture (10). EMDEP validates data via an interactive depositor-driven operation, relying on the knowledge and expertise of the experimenters for the complete and accurate description of the structural experiment and its results. The captured metadata (e.g. sample description, specimen preparation, imaging, reconstruction and fitting details) are stored in an XML-style ‘header’ file, and the deposited map is converted to a common format for redistribution. A database query tool, EMSEARCH, was also designed and implemented to enable web-based searches. By December 2003, the EMDB was an operational public database with 65 maps deposited by major EM laboratories from Europe and the USA. At this time the PDB began to see an increase in EM-related coordinate depositions, in many cases models fitted into maps deposited to EMDB.
Models
The PDB archive was established in 1971 as a public repository for X-ray crystal structures of biological macromolecules (11), and is presently maintained by the global organization world-wide PDB [wwPDB (12)]. The number of structures in the PDB has grown from the initial 7 to over 67 000 entries. Over time, the PDB began to collect coordinates of structures determined by methods other than X-ray crystallography, including NMR spectroscopy, neutron diffraction, fiber diffraction, electron crystallography, electron microscopy and solution scattering. Coordinates for structures determined using EM began to be archived in the 1990s, beginning with models for bacteriorhodopsin (13) and the RecA hexamer (14). Currently, the deposition rate for EM entries in the PDB is increasing more rapidly than for any other experimental method.
EM dictionary development
Two workshops held in 2004 (3DEM NoE workshop at EBI and CryoEM Structure Deposition Workshop at RCSB-PDB, co-sponsored by NCMI) invited the EM community to participate in development of an improved data model for describing cryoEM experiments, and also set in motion efforts to unify deposition and access to EM-derived maps and models. Following the workshops, a revised and expanded EM dictionary handling both map and model metadata was created in a three-way collaboration between PDBe, RCSB and NCMI with broad community input and was presented at the 2005 3DEM Gordon Research Conference in New Hampshire as well as a 3DEM Developers workshop held in the UK. During this period, Heymann et al. (15) also published recommended conventions for exchange of cryoEM data, and a standards task force was created to gather information on the different cryoEM map and image conventions and formats to facilitate conversion.
Unified resource
Following the recommendations of the EM community to create a ‘one-stop shop for EM,’ the Unified Data Resource for CryoEM (EMDataBank.org) was established in 2007 with funding from the National Institutes of Health/NIGMS as a joint effort of PDBe, RCSB and NCMI. The mission of the resource is to build up a global deposition and retrieval network for cryoEM map, model and associated metadata, as well as a portal for software tools for standardized map format conversion, map, segmentation and model assessment, visualization and data integration. The first goal of this collaboration was achieved in early 2008 when RCSB joined PDBe as a second map deposition and retrieval site. Joint EMDB (map) and PDB (model) deposition systems were developed and put into operation at both PDBe and RCSB in early 2009, and web-based 3D visualization tools have been integrated into EMDB atlas pages. Efforts to improve uniformity and usability of the EM structural data in both EMDB and PDB databases are ongoing; we recently completed remediation of voxel sizes and density statistics stored in all EMDB map files to ensure display at correct physical scale and with reasonable density contour level. Additional services for data harvesting and evaluation are planned.
DEPOSITION AND CONTENT
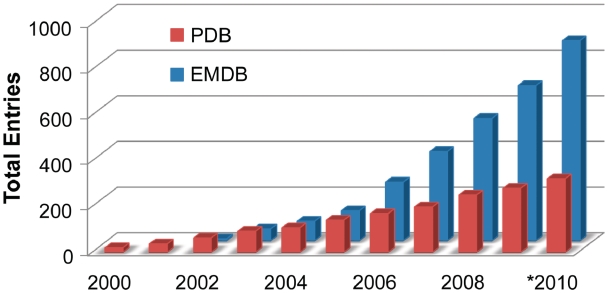
The EMDB currently holds more than 800 map entries with resolution limits ranging from 80 to 2 Å, while PDB holds more than 300 entries of coordinates fitted into EM map volumes (Figure 1). Map volume and fitted model deposition rates in 2008–09 were ∼150 and ∼40 per year, respectively, with roughly 40% of all published EM structures being captured in the databases. As the importance of EM-derived structural information continues to increase, it is anticipated that more journals and funding agencies will require deposition to structural databases.
Figure 1.
EM entries in EMDB and PDB, cumulative by year (*2010 data is through July).
Each EMDB entry holds a single map plus associated experimental metadata; each PDB entry holds a fitted coordinate model and associated experimental metadata plus primary sequence information for each polymer. The metadata information that is shared by both databases is automatically transferred during the joint deposition process and includes:
sample description, including components and stoichiometry;
sample and specimen preparation, including buffer, grid, vitrification, staining;
imaging experiment, including microscope type, microscope settings, detector;
specimen selection, image correction, reconstruction method, resolution/resolution method; and
fitting description, including PDB ids of source coordinates.
Correspondences between maps and associated fitted coordinate models are maintained within both archives. EMDB entries can optionally hold associated masks, structure factors and/or layerline data; PDB entries can also hold structure factors. The underlying dictionaries for the two databases have direct translations and are regularly updated to reflect changes in experimental apparatus and methods.
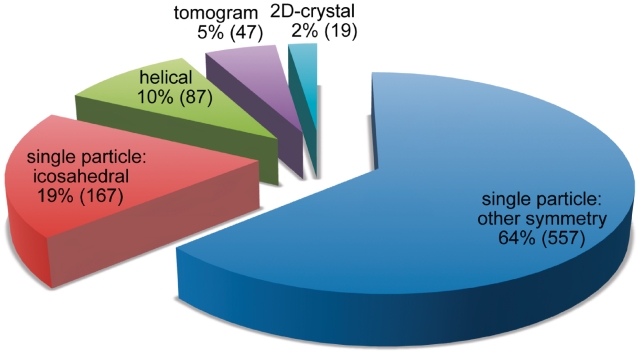
The map archive includes maps generated by a number of different electron microscopy reconstruction methods (Figure 2). The majority of entries (83%) are single particle reconstructions, which represent ensemble averages of thousands of individual imaged particles, often with additional symmetry averaging. The largest class of single particle specimens represented are the viruses (20% of all holdings), the majority of which are icosahedrally averaged. Virus entries typically represent distinct states of maturation, or complexes with antibodies or receptors. The second largest class of single particle specimens represented are the ribosomes (15%). Ribosome entries define key structural conformations encountered in translating messenger RNA into protein, or elucidate structural variations across diverse species. Other single particle specimens represented include macromolecular machines involved in protein folding, protein degradation, energy metabolism, cell cycle processes, DNA replication, DNA repair, RNA transcription, RNA splicing and ion channels.
Figure 2.
Current distribution of EM map entry types.
The map archive also holds densities for 2D crystals and for helical arrays, including intracellular filaments and microtubules, flagella and helical crystals. There are also several tomographic maps of unique structures as well as maps that represent 3D averages of aligned tomograms. Diverse specimens currently held of this type include flagellar motors, insect flight muscle tissue, desmosomes and viruses.
EM-derived coordinates are obtained using a variety of modeling methods including manual docking, rigid-body fitting, homology modeling, de novo modeling and computational refinement algorithms. EM entries in the PDB are classified either under electron microscopy or electron crystallography as the experimental method. For structures with regular point or helical symmetry, coordinates are given for the asymmetric unit along with a set of transformation matrices to build the biological assembly (16).
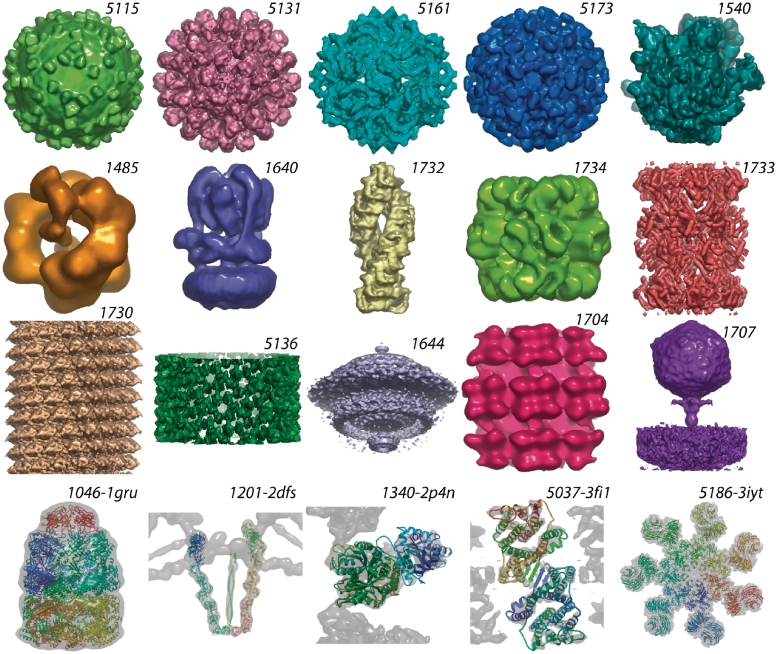
A gallery of representative maps including some maps with fitted models is presented in Figure 3.
Figure 3.
Image gallery of EM structures in EMDB and PDB (labels are database ids with “EMD-” prefixes omitted for brevity; scaling is arbitrary). Icosahedral—5115: West Nile virus-antibody complex (53); 5131: wild-rabbit hemorrhagic disease virus (54); 5161: Partitivirus (55); 5173: Hepatitis E particles (56); other single particle: 1540: pre-translocational ribosome (57); 1485: DNA ligase–PCNA–DNA complex (58); 1640: yeast V-ATPase (59); 1732: tripeptidyl peptidase II (60); 1734: rubisco-methyltransferase complex (61); 1733: 20S proteasome (62); Helical—1730: tobacco mosaic virus (63); 5136: HIV-1 capsid tubular crystal (64); Tomogram averages—1644: Borrelia flagellar motor (65); 1704: Tula Hantavirus glycoprotein spikes (66); 1707: Podovirus P-SSP7 infecting Prochlorococcus marinus (67); Map+model pairs—1046-1gru: GroES–GroEL complex (68); 1201-2dfs: Myosin V (69); 1340-2p4n: kinesin bound to a microtubule (70); 5037-3fi1: ion-coupled transport protein NhaA (71); 5186-3iyt: apoptosome–procaspase-9 CARD complex (72). All images were created using OpenAstexViewer as implemented on the EMDB atlas pages for these entries.
ACCESS
Access to EM structural data and related services is through the EMDataBank.org web site (Table 1). The EMDB and PDB archives are updated weekly on Wednesdays at 00:00 GMT. EMDB is distributed on ftp mirrors supported by the two EMDataBank.org distribution partners in the UK and the USA, while PDB is distributed on ftp mirrors supported by each of the wwPDB partners. Upon depositor request, EM entries may be held for up to 2 years from the deposition date for map entries in EMDB, and up to 1 year for coordinate models in PDB. However, we strongly encourage depositors to make their data publicly available as soon as possible.
Table 1.
Overview of EMDataBank.org resources and services as of September 2010
| EMDataBank.org page | Description |
|---|---|
| /deposit.html | Links to EMDEP deposition servers hosted by RCSB-PDB and PDBe, CryoEM Map+Model Deposition Guide and EMDB Hold/Release Policy |
| /search.html | Links to EMSEARCH servers hosted by RCSB-PDB and PDBe, Full Text Search (α) |
| /recententries.html | Table of all recently released and recently submitted EMDB entries, updated weekly |
| /webservice.html | EMDataBank web service based on the SOAP protocol with application components for use by other applications over the Internet |
| /otherservices.html | Links to other services including the ftp servers at PDBe (ftp://ftp.ebi.ac.uk/pub/databases/emdb) and RCSB-PDB (ftp://emdb.rutgers.edu) |
| /allnews.html | EMDataBank.org news items |
| /faq.html | Frequently asked questions about EM map volume depositions, EM fitted coordinate model depositions and map access |
| /mapformat.html | EMDB map distribution format description (CCP4 format for 3DEM maps) |
| /emsoftware.html | List of software available within the EM community for generation, analysis and fitting of EM maps, sortable by program name and type |
| /emtestdata.html | Links to EM image data sets that have been made publicly available by EM community members to download for testing |
The EMSEARCH web service is maintained and updated weekly at both distribution sites. EMSEARCH enables browsing and searching of EMDB metadata uploaded into a relational database. Simple searches can be performed based on author name, title, entry id, sample name, citation abstract word, aggregation type, resolution and release date range. Search summaries link to a set of atlas pages for each entry, which include ‘Summary’, ‘Visualization’, ‘Sample’, ‘Experiment’, ‘Processing’, ‘Map Information’ and ‘Download’ pages (Figure 4). In addition, a full-text search system based on the Lucene indexer is nearing completion. All search options can be accessed from the EMDataBank.org search tab.
Figure 4.
Example EMDB atlas page.
The ‘Visualization’ page for each EM map entry provides several viewing options. In addition to a static 2D image provided by the deposition author, it is also possible to launch two different Java-based 3D viewers (Figure 5, left and right top panels). EMViewer provides a simple, single isosurface representation of a map at a predetermined contour level that can be manipulated by mouse drag and click actions. OpenAstexViewer, a molecular graphics program originally developed to display crystallographic data (17) has been adapted for display of EM maps and associated coordinate models. To improve web download speed and minimize memory requirements, a compact map format is used (BRIX), and larger maps are also down-sampled by a factor of 2–5. Current capabilities include ability to control map contour level, opacity, color, solid versus mesh surface rendering and concurrent display of a PDB coordinate entry. Viewing large maps may require increasing Java Applet Runtime memory allocation.
Figure 5.
Tools for 3D visualization of EM structural data. EM viewer (top left) and OpenAstexViewer (top right) are Java-based 3D viewers that can be launched from the EMDB atlas ‘Visualization’ pages. EM viewer employs a compact single contour-level mesh representation for lightweight map viewing; OpenAstexViewer enables exploration of maps and their fitted models with adjustable map contour level. The EMDataBank.org web service implemented in UCSF chimera (bottom panel) expedites search of the EMDB and subsequent map + model download. The example shown is GroEL [EMD-5001and PDB 3CAU; (73)].
Maps are distributed in CCP4 format and can be viewed with locally installed software such as UCSF Chimera (18), Pymol (19), VMD (20), Coot (21), enabling further analysis and manipulation with an extensive set of tools. Links for map download are available on the ‘Download’ and ‘Map Information’ pages of each entry. Recent distributions of UCSF Chimera enable direct downloads of EMDB maps plus associated PDB models via simple queries (e.g. author name, title) to the EMSEARCH relational database through the EMDataBank.org beta-web service, a self-contained programing interface based on the SOAP protocol (Figure 5, bottom panel).
EXAMPLES OF USE
A major goal of the EMDataBank.org unified data resource is to archive EM-derived structural information in a way that will enable further research. The impact of availability of structural data for smaller proteins has already been amply demonstrated by the success of the PDB, which is accessed by thousands of individuals around the globe every day. Listed here are a few examples of how archived EM data facilitates subsequent scientific exploration by other investigators.
Building up molecular pictures of large macromolecular assemblies
Atomic coordinates permit 3D mapping of a wide variety of data including sites of mutations leading to disease, modification sites, antibody recognition sites, amino acid sequence variability and electrostatic properties. But in many cases, high-resolution structural models are not available for every component within a cryoEM map at the time it is first interpreted. By preserving map and model information together in a freely accessible database, new structural information can be incorporated as it becomes available. Examples include (i) reinterpretation of ribosome stalk regions in multiple archived maps [EMD-1005 through 1008, 1055, 1056; (22)] and (ii) recent progress by the Rossmann group towards complete fitted coordinate models for bacteriophage T4 in multiple functional states [EMD-1048 and 3H3W; EMD-1086 and 3H3Y (23)].
Structural basis for interpreting data
Structural knowledge can be crucial for interpreting biochemical and biophysical data. For instance, dengue and West Nile virus cryoEM structures (24,25) have led the way to mapping sites of glycosylation (26), amino acid sequence variability (27), neutralizing antibody binding (28), design of vaccines (29) and a structural basis for understanding membrane fusion (30). Fitting of X-ray crystal structures of cadherin into tomographic maps of epidermal desmosomes have yielded insights into cell adhesion [EMD-1051, 1052, 1053, 1374, 1449; (31,32)].
Crystal structure phasing
For low-symmetry complexes such as ribosomes, a low-resolution cryoEM map can provide a valuable complementary source of information for crystallographic phasing. For high-symmetry structures such as icosahedral viruses, application of robust computational averaging and extension algorithms to initial cryoEM-derived phases is often sufficient to complete the structure determination without additional phase information. Recent case studies have employed ribosome maps [EMD-1008 and 1019; (33)] and a dodecameric enzyme map [EMD-1680; (34)]. Routine archiving of EM-derived maps facilitates the use of this method by the crystallography community.
Comparative studies
By capturing coordinates, maps and metadata and making them available through searchable databases, it is possible to compare and select structures for further analysis using a variety of criteria, perform experimental design for new analyses, and design new algorithms to improve the state of the art methodology. For example, comparison of publicly available maps and coordinates led to the conclusion that the double-stranded DNA tailed phages and herpesvirus have a similar fold in their major capsid proteins though there is little sequence similarity among them [EMD-1101 and PDB entries 1OHG, 1YUE; (35)]. In a second example, a flexible fitting approach applied to 43 maps of bacterial 70S ribosome from EMDB in various functional states revealed global conformational differences between the EM structures involving large-scale ratchet-like deformations (36).
Structures not accessible by other techniques
Many large molecular machines are not amenable to crystallization for high-resolution studies. CryoEM methods have begun to yield structures of membrane proteins, viruses and chaperonins determined to resolution limits beyond 4.5 Å where de novo methods can trace a Cα-backbone. In many cases, side chain densities are also visible. In one recent example, over 80% of side-chain densities in the archaeal chaperonin hexadecamer Mm-cpn chaperonin could be resolved [EMD-5137 and PDB 3LOS; (37)].
Software development
Availability of EM map volumes in the EMDB facilitates development and validation of software for map viewing, analysis, manipulation, coordinate model fitting and validation. Development of algorithms for fitting of EM maps with atomic coordinates is a particularly active area. For low to medium-resolution studies in which the source coordinates are likely to be derived from crystallographic or NMR studies, algorithms being explored include rigid-body fitting (38), normal mode analysis (39), spatial interpolation (40), conformational sampling under low-resolution restraints (37,41), molecular dynamics flexible fitting (42) and simulated annealing approaches (43,44). For medium- to high-resolution studies where de novo model building becomes possible, methods are being developed for skeletonization and secondary-structure element detection (45) and incorporation of structure prediction from primary sequence (46). Additional examples of algorithm development facilitated by public availability of EMDB maps include investigations of map denoising (47), B-factor sharpening (48), map resolution determination (49) and automated segmentation (50,51). The UCSF Chimera team has made particularly effective use of the EMDB resource for development and testing of a large, versatile set of tools for manipulating volume data (18,52).
FUTURE PROSPECTS
3D cryoEM reconstruction methods are uniquely able to reveal structural aspects of many important macromolecules and macromolecular complexes, and for this reason the field is in a period of rapid expansion and development. Based on current growth of EM entries and publication of EM structures, the total number of structures of large biological assemblies contributed by EM is anticipated to approach 10 000 by the year 2020.
In the near future, deposition and archiving of EM structural data will be integrated in a common tool that is being developed by the wwPDB partners to handle depositions from all structural biology methods. In addition, as the field matures, validation tools and criteria for assessment of map and fitted coordinate models will play an important role in providing guidance to users of cryoEM derived structural data. To this end, a validation task force is being assembled by the unified data resource partners (NCMI, RCSB and PDBe) to develop recommendations as to how best to assess the quality of both maps and models that have been obtained from cryoEM data. The recommendations will form the basis for a validation suite to be used by EMDB and PDB. The ‘CryoEM Modelling Challenge 2010′ (http://ncmi.bcm.edu/challenge), in which the modeling community is being asked to apply their modeling tools to a selected set of cryoEM densities at different resolutions, will permit careful comparison of the many modeling methods under development and will yield useful benchmark data for validation suite development.
FUNDING
EMDataBank.org is supported by a National Institutes of Health grant [R01GM079429] to Baylor College of Medicine, Rutgers University and the European Bioinformatics Institute. PDBe receives additional support for EMDB development from the Biotechnology and Biological Sciences Research Council [BBG022577] and from EMBL. Funding for open access charge: NIH grant R01GM079429.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We are grateful to Tom Goddard for integrating the EMDataBank.org web service into UCSF Chimera.
REFERENCES
- 1.Frank J. Three-dimensional Electron Microscopy of Macromolecular Assemblies: Visualization of Biological Molecules in their Native State. New York: Oxford University Press; 2006. [Google Scholar]
- 2.Glaeser RM, Downing KH, DeRosier D, Chiu W, Frank J. Electron Crystallography of Biological Macromolecules. Oxford, New York: Oxford University Press; 2007. [Google Scholar]
- 3.Chiu W, Baker ML, Jiang W, Dougherty M, Schmid MF. Electron cryomicroscopy of biological machines at subnanometer resolution. Structure. 2005;13:363–372. doi: 10.1016/j.str.2004.12.016. [DOI] [PubMed] [Google Scholar]
- 4.Rossmann MG, Morais MC, Leiman PG, Zhang W. Combining x-ray crystallography and electron microscopy. Structure. 2005;13:355–362. doi: 10.1016/j.str.2005.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Baker ML, Baker MR, Hryc CF, Dimaio F. Analyses of subnanometer resolution cryoEM density maps. In: Jensen G, editor. Methods in Enzymology. Vol. 483. London: Elsevier; 2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tagari M, Newman R, Chagoyen M, Carazo JM, Henrick K. New electron microscopy database and deposition system. Trends Biochem. Sci. 2002;27:589. doi: 10.1016/s0968-0004(02)02176-x. [DOI] [PubMed] [Google Scholar]
- 7.Dutta S, Burkhardt K, Young J, Swaminathan GJ, Matsuura T, Henrick K, Nakamura H, Berman HM. Data deposition and annotation at the worldwide protein data bank. Mol. Biotechnol. 2009;42:1–13. doi: 10.1007/s12033-008-9127-7. [DOI] [PubMed] [Google Scholar]
- 8.Fuller SD. Depositing electron microscopy maps. Structure. 2003;11:11–12. doi: 10.1016/s0969-2126(02)00942-5. [DOI] [PubMed] [Google Scholar]
- 9.Editorial. Nat. Struct. Biol. 2003;10:313. doi: 10.1038/nsb0503-313. [DOI] [PubMed] [Google Scholar]
- 10.Henrick K, Newman R, Tagari M, Chagoyen M. EMDep: a web-based system for the deposition and validation of high-resolution electron microscopy macromolecular structural information. J. Struct. Biol. 2003;144:228–237. doi: 10.1016/j.jsb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Bernstein FC, Koetzle TF, Williams GJB, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 12.Berman HM, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 13.Henderson R, Baldwin JM, Ceska TA, Zemlin F, Beckmann E, Downing KH. Model for the structure of bacteriorhodopsin based on high-resolution electron cryo-microscopy. J. Mol. Biol. 1990;213:899–929. doi: 10.1016/S0022-2836(05)80271-2. [DOI] [PubMed] [Google Scholar]
- 14.Yu X, Egelman EH. The RecA hexamer is a structural homologue of ring helicases. Nat. Struct. Biol. 1997;4:101–104. doi: 10.1038/nsb0297-101. [DOI] [PubMed] [Google Scholar]
- 15.Heymann JB, Chagoyen M, Belnap DM. Common conventions for interchange and archiving of three-dimensional electron microscopy information in structural biology. J. Struct. Biol. 2005;151:196–207. doi: 10.1016/j.jsb.2005.06.001. [DOI] [PubMed] [Google Scholar]
- 16.Lawson CL, Dutta S, Westbrook JD, Henrick K, Berman HM. Representation of viruses in the remediated PDB archive. Acta Crystallogr. D Biol. Crystallogr. 2008;64:874–882. doi: 10.1107/S0907444908017393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartshorn MJ. AstexViewer: a visualisation aid for structure-based drug design. J. Comput. Aided Mol. Des. 2002;16:871–881. doi: 10.1023/a:1023813504011. [DOI] [PubMed] [Google Scholar]
- 18.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera–a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 19.Delano WL. The Pymol Molecular Graphics System. Palo Alto: Delano Scientific; 2002. [Google Scholar]
- 20.Hsin J, Arkhipov A, Yin Y, Stone JE, Schulten K. Using VMD: an introductory tutorial. Curr. Protoc. Bioinformatics. 2008 doi: 10.1002/0471250953.bi0507s24. Chapter 5, Unit 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Emsley P, Cowtan K. Coot: model-building tools for molecular graphics. Acta Crystallogr. D Biol. Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 22.Diaconu M, Kothe U, Schlunzen F, Fischer N, Harms JM, Tonevitsky AG, Stark H, Rodnina MV, Wahl MC. Structural basis for the function of the ribosomal L7/12 stalk in factor binding and GTPase activation. Cell. 2005;121:991–1004. doi: 10.1016/j.cell.2005.04.015. [DOI] [PubMed] [Google Scholar]
- 23.Aksyuk AA, Leiman PG, Shneider MM, Mesyanzhinov VV, Rossmann MG. The structure of gene product 6 of bacteriophage T4, the hinge-pin of the baseplate. Structure. 2009;17:800–808. doi: 10.1016/j.str.2009.04.005. [DOI] [PubMed] [Google Scholar]
- 24.Kuhn RJ, Zhang W, Rossmann MG, Pletnev SV, Corver J, Lenches E, Jones CT, Mukhopadhyay S, Chipman PR, Strauss EG, et al. Structure of dengue virus: implications for flavivirus organization, maturation, and fusion. Cell. 2002;108:717–725. doi: 10.1016/s0092-8674(02)00660-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mukhopadhyay S, Kim BS, Chipman PR, Rossmann MG, Kuhn RJ. Structure of West Nile virus. Science. 2003;302:248. doi: 10.1126/science.1089316. [DOI] [PubMed] [Google Scholar]
- 26.Hanna SL, Pierson TC, Sanchez MD, Ahmed AA, Murtadha MM, Doms RW. N-linked glycosylation of west nile virus envelope proteins influences particle assembly and infectivity. J. Virol. 2005;79:13262–13274. doi: 10.1128/JVI.79.21.13262-13274.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Modis Y, Ogata S, Clements D, Harrison SC. Variable surface epitopes in the crystal structure of dengue virus type 3 envelope glycoprotein. J. Virol. 2005;79:1223–1231. doi: 10.1128/JVI.79.2.1223-1231.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Nybakken GE, Oliphant T, Johnson S, Burke S, Diamond MS, Fremont DH. Structural basis of West Nile virus neutralization by a therapeutic antibody. Nature. 2005;437:764–768. doi: 10.1038/nature03956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ledizet M, Kar K, Foellmer HG, Tian W, Bushmich SL, Anderson JF, Fikrig E, Koski RA. A recombinant envelope protein vaccine against West Nile virus. Vaccine. 2005;23:3915–3924. doi: 10.1016/j.vaccine.2005.03.006. [DOI] [PubMed] [Google Scholar]
- 30.Modis Y, Ogata S, Clements D, Harrison SC. Structure of the dengue virus envelope protein after membrane fusion. Nature. 2004;427:313–319. doi: 10.1038/nature02165. [DOI] [PubMed] [Google Scholar]
- 31.He W, Cowin P, Stokes DL. Untangling desmosomal knots with electron tomography. Science. 2003;302:109–113. doi: 10.1126/science.1086957. [DOI] [PubMed] [Google Scholar]
- 32.Al-Amoudi A, Diez DC, Betts MJ, Frangakis AS. The molecular architecture of cadherins in native epidermal desmosomes. Nature. 2007;450:832–837. doi: 10.1038/nature05994. [DOI] [PubMed] [Google Scholar]
- 33.Xiong Y. From electron microscopy to X-ray crystallography: molecular-replacement case studies. Acta Crystallogr. D Biol. Crystallogr. 2008;64:76–82. doi: 10.1107/S090744490705398X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Trapani S, Schoehn G, Navaza J, Abergel C. Macromolecular crystal data phased by negative-stained electron-microscopy reconstructions. Acta Cryst. D. 2010;66:514–521. doi: 10.1107/S0907444910002763. [DOI] [PubMed] [Google Scholar]
- 35.Baker ML, Jiang W, Rixon FJ, Chiu W. Common ancestry of herpesviruses and tailed DNA bacteriophages. J. Virol. 2005;79:14967–14970. doi: 10.1128/JVI.79.23.14967-14970.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto A, Ishida H. Global conformational changes of ribosome observed by normal mode fitting for 3D CryoEM structures. Structure. 2009;17:1605–1613. doi: 10.1016/j.str.2009.09.017. [DOI] [PubMed] [Google Scholar]
- 37.Zhang J, Baker ML, Schroder GF, Douglas NR, Reissmann S, Jakana J, Dougherty M, Fu CJ, Levitt M, Ludtke SJ, et al. Mechanism of folding chamber closure in a group II chaperonin. Nature. 2010;463:379–383. doi: 10.1038/nature08701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wriggers W. Using Situs for the integration of multi-resolution structures. Biophys. Rev. 2010;2:21–27. doi: 10.1007/s12551-009-0026-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tama F, Miyashita O, Brooks CL., 3rd Normal mode based flexible fitting of high-resolution structure into low-resolution experimental data from cryoEM. J. Struct. Biol. 2004;147:315–326. doi: 10.1016/j.jsb.2004.03.002. [DOI] [PubMed] [Google Scholar]
- 40.Rusu M, Birmanns S, Wriggers W. Biomolecular pleiomorphism probed by spatial interpolation of coarse models. Bioinformatics. 2008;24:2460–2466. doi: 10.1093/bioinformatics/btn461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schroder GF, Brunger AT, Levitt M. Combining efficient conformational sampling with a deformable elastic network model facilitates structure refinement at low resolution. Structure. 2007;15:1630–1641. doi: 10.1016/j.str.2007.09.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Trabuco LG, Villa E, Schreiner E, Harrison CB, Schulten K. Molecular dynamics flexible fitting: a practical guide to combine cryo-electron microscopy and X-ray crystallography. Methods. 2009;49:174–180. doi: 10.1016/j.ymeth.2009.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tan RK, Devkota B, Harvey SC. YUP.SCX: coaxing atomic models into medium resolution electron density maps. J. Struct. Biol. 2008;163:163–174. doi: 10.1016/j.jsb.2008.05.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Topf M, Lasker K, Webb B, Wolfson H, Chiu W, Sali A. Protein structure fitting and refinement guided by cryoEM density. Structure. 2008;16:295–307. doi: 10.1016/j.str.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Baker ML, Ju T, Chiu W. Identification of secondary structure elements in intermediate-resolution density maps. Structure. 2007;15:7–19. doi: 10.1016/j.str.2006.11.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.DiMaio F, Tyka MD, Baker ML, Chiu W, Baker D. Refinement of protein structures into low-resolution density maps using Rosetta. J. Mol. Biol. 2009;392:181–190. doi: 10.1016/j.jmb.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Jiang W, Baker M, Wu Q, Bajaj C, Chiu W. Applications of bilateral denoising filter in biological electron microscopy. J. Struct. Biol. 2003;144:114–122. doi: 10.1016/j.jsb.2003.09.028. [DOI] [PubMed] [Google Scholar]
- 48.Fernandez JJ, Luque D, Caston JR, Carrascosa JL. Sharpening high resolution information in single particle electron cryomicroscopy. J. Struct. Biol. 2008;164:170–175. doi: 10.1016/j.jsb.2008.05.010. [DOI] [PubMed] [Google Scholar]
- 49.Sousa D, Grigorieff N. Ab initio resolution measurement for single particle structures. J. Struct. Biol. 2007;157:201–210. doi: 10.1016/j.jsb.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 50.Baker ML, Yu Z, Chiu W, Bajaj C. Automated segmentation of molecular subunits in electron cryomicroscopy density maps. J. Struct. Biol. 2006;156:432–441. doi: 10.1016/j.jsb.2006.05.013. [DOI] [PubMed] [Google Scholar]
- 51.Pintilie GD, Zhang J, Goddard TD, Chiu W, Gossard DC. Quantitative analysis of cryoEM density map segmentation by watershed and scale-space filtering, and fitting of structures by alignment to regions. J. Struct. Biol. 2010;170:427–438. doi: 10.1016/j.jsb.2010.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Goddard TD, Huang CC, Ferrin TE. Visualizing density maps with UCSF Chimera. J. Struct. Biol. 2007;157:281–287. doi: 10.1016/j.jsb.2006.06.010. [DOI] [PubMed] [Google Scholar]
- 53.Kaufmann B, Chipman PR, Holdaway HA, Johnson S, Fremont DH, Kuhn RJ, Diamond MS, Rossmann MG. Capturing a flavivirus pre-fusion intermediate. PLoS Pathog. 2009;5:e1000672. doi: 10.1371/journal.ppat.1000672. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hu Z, Tian X, Zhai Y, Xu W, Zheng D, Sun F. Cryo-electron microscopy reconstructions of two types of wild rabbit hemorrahagic disease viruses characterized the structural features of Lagovirus. Protein Cell. 2010;1:48–58. doi: 10.1007/s13238-010-0007-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tang J, Pan J, Havens WM, Ochoa WF, Guu TS, Ghabrial SA, Nibert ML, Tao YJ, Baker TS. Backbone trace of partitivirus capsid protein from electron cryomicroscopy and homology modeling. Biophys. J. 2010;99:685–694. doi: 10.1016/j.bpj.2010.04.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Xing L, Li TC, Miyazaki N, Simon MN, Wall JS, Moore M, Wang CY, Takeda N, Wakita T, Miyamura T, et al. Structure of hepatitis E virion-sized particle reveals an RNA-dependent viral assembly pathway. J. Biol. Chem. 2010;285:33175–33183. doi: 10.1074/jbc.M110.106336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Agirrezabala X, Lei J, Brunelle JL, Ortiz-Meoz RF, Green R, Frank J. Visualization of the hybrid state of tRNA binding promoted by spontaneous ratcheting of the ribosome. Mol. Cell. 2008;32:190–197. doi: 10.1016/j.molcel.2008.10.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Mayanagi K, Kiyonari S, Saito M, Shirai T, Ishino Y, Morikawa K. Mechanism of replication machinery assembly as revealed by the DNA ligase-PCNA-DNA complex architecture. Proc. Natl Acad. Sci. USA. 2009;106:4647–4652. doi: 10.1073/pnas.0811196106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Diepholz M, Venzke D, Prinz S, Batisse C, Florchinger B, Rossle M, Svergun DI, Bottcher B, Fethiere J. A different conformation for EGC stator subcomplex in solution and in the assembled yeast V-ATPase: possible implications for regulatory disassembly. Structure. 2008;16:1789–1798. doi: 10.1016/j.str.2008.09.010. [DOI] [PubMed] [Google Scholar]
- 60.Chuang CK, Rockel B, Seyit G, Walian PJ, Schonegge AM, Peters J, Zwart PH, Baumeister W, Jap BK. Hybrid molecular structure of the giant protease tripeptidyl peptidase II. Nat. Struct. Mol. Biol. 2010;17:990–996. doi: 10.1038/nsmb.1870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Raunser S, Magnani R, Huang Z, Houtz RL, Trievel RC, Penczek PA, Walz T. Rubisco in complex with Rubisco large subunit methyltransferase. Proc. Natl Acad. Sci. USA. 2009;106:3160–3165. doi: 10.1073/pnas.0810563106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rabl J, Smith DM, Yu Y, Chang SC, Goldberg AL, Cheng Y. Mechanism of gate opening in the 20S proteasome by the proteasomal ATPases. Mol. Cell. 2008;30:360–368. doi: 10.1016/j.molcel.2008.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clare DK, Orlova EV. 4.6A CryoEM reconstruction of tobacco mosaic virus from images recorded at 300 keV on a 4k x 4k CCD camera. J. Struct. Biol. 2010;171:303–308. doi: 10.1016/j.jsb.2010.06.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Byeon IJ, Meng X, Jung J, Zhao G, Yang R, Ahn J, Shi J, Concel J, Aiken C, Zhang P, et al. Structural convergence between CryoEM and NMR reveals intersubunit interactions critical for HIV-1 capsid function. Cell. 2009;139:780–790. doi: 10.1016/j.cell.2009.10.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kudryashev M, Cyrklaff M, Wallich R, Baumeister W, Frischknecht F. Distinct in situ structures of the Borrelia flagellar motor. J. Struct. Biol. 2010;169:54–61. doi: 10.1016/j.jsb.2009.08.008. [DOI] [PubMed] [Google Scholar]
- 66.Huiskonen JT, Hepojoki J, Laurinmaki P, Vaheri A, Lankinen H, Butcher SJ, Grunewald K. Electron cryotomography of Tula hantavirus suggests a unique assembly paradigm for enveloped viruses. J. Virol. 2010;84:4889–4897. doi: 10.1128/JVI.00057-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Liu X, Zhang Q, Murata K, Baker ML, Sullivan MB, Fu C, Dougherty MT, Schmid MF, Osburne MS, Chisholm SW, et al. Structural changes in a marine podovirus associated with release of its genome into Prochlorococcus. Nat. Struct. Mol. Biol. 2010;17:830–836. doi: 10.1038/nsmb.1823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Ranson NA, Farr GW, Roseman AM, Gowen B, Fenton WA, Horwich AL, Saibil HR. ATP-bound states of GroEL captured by cryo-electron microscopy. Cell. 2001;107:869–879. doi: 10.1016/s0092-8674(01)00617-1. [DOI] [PubMed] [Google Scholar]
- 69.Liu J, Taylor DW, Krementsova EB, Trybus KM, Taylor KA. Three-dimensional structure of the myosin V inhibited state by cryoelectron tomography. Nature. 2006;442:208–211. doi: 10.1038/nature04719. [DOI] [PubMed] [Google Scholar]
- 70.Sindelar CV, Downing KH. The beginning of kinesin's force-generating cycle visualized at 9-A resolution. J. Cell Biol. 2007;177:377–385. doi: 10.1083/jcb.200612090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Williams KA. Three-dimensional structure of the ion-coupled transport protein NhaA. Nature. 2000;403:112–115. doi: 10.1038/47534. [DOI] [PubMed] [Google Scholar]
- 72.Yuan S, Yu X, Topf M, Ludtke SJ, Wang X, Akey CW. Structure of an apoptosome-procaspase-9 CARD complex. Structure. 2010;18:571–583. doi: 10.1016/j.str.2010.04.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Ludtke SJ, Baker ML, Chen DH, Song JL, Chuang DT, Chiu W. De novo backbone trace of GroEL from single particle electron cryomicroscopy. Structure. 2008;16:441–448. doi: 10.1016/j.str.2008.02.007. [DOI] [PubMed] [Google Scholar]