Abstract
DBASS3 and DBASS5 provide comprehensive repositories of new exon boundaries that were induced by pathogenic mutations in human disease genes. Aberrant 5′- and 3′-splice sites were activated either by mutations in the consensus sequences of natural exon–intron junctions (cryptic sites) or elsewhere (‘de novo’ sites). DBASS3 and DBASS5 currently contain approximately 900 records of cryptic and de novo 3′- and 5′-splice sites that were produced by over a thousand different mutations in approximately 360 genes. DBASS3 and DBASS5 data can be searched by disease phenotype, gene, mutation, location of aberrant splice sites in introns and exons and their distance from authentic counterparts, by bibliographic references and by the splice-site strength estimated with several prediction algorithms. The user can also retrieve reference sequences of both aberrant and authentic splice sites with the underlying mutation. These data will facilitate identification of introns or exons frequently involved in aberrant splicing, mutation analysis of human disease genes and study of germline or somatic mutations that impair RNA processing. Finally, this resource will be useful for fine-tuning splice-site prediction algorithms, better definition of auxiliary splicing signals and design of new reporter assays. DBASS3 and DBASS5 are freely available at http://www.dbass.org.uk/.
INTRODUCTION
Pre-mRNA splicing removes intervening sequences or introns from eukaryotic precursor messenger RNAs (pre-mRNAs) to ensure accurate gene expression (1). Apart from joining consecutive exons together, this process is capable of selective removal or inclusion of exonic and intronic segments in mRNA, generating distinct transcripts from a single gene, often in a cell type-specific or developmental- or gender-dependent manner (2–4). Both constitutive and alternative splicing are controlled by sequence elements in the pre-mRNA that are recognized by a large ribonucleoprotein complex termed the spliceosome (1). These conserved but degenerate signals are located predominantly in introns and include 5′-splice sites (5′-ss) and 3′-splice sites (3′-ss), with upstream polypyrimidine tracts and the branch point sequence. Mutations in any of these cis-elements can dramatically alter splicing efficiency and result in genetic disease (5–8), but their consequences for RNA processing have been difficult to predict.
The majority of mutations at the 5′-ss or 3′-ss consensus have been reported to cause skipping of one or more exons and activation of cryptic splice sites (9). Occasionally, splice-site mutations may also result in full intron retention or give rise to entirely novel ‘pseudoexons’ or cryptic exons, often in repetitive sequences, particularly in short interspersed nuclear elements (SINEs), including mammalian interspersed repeats and Alus (10–12). Finally, creation of de novo splice sites in large exons may remove internal exonic sequences and create pseudointrons.
Traditional splicing signals contain only a half of the information necessary for accurate splice site recognition (13). The remaining information is provided by auxiliary signals in introns and exons, known as splicing enhancers and silencers (14,15), that are thought to interact with trans-acting factors and/or contribute to critical RNA structural motifs and a ‘splicing code’ (16,17). In addition, as the splicing and transcription machineries are tightly linked, splicing outcomes can be influenced by pre-mRNA processing kinetics and transcription (18). As a result, a ‘splicing mutation’ may affect not only RNA processing, but also transcription (19) and downstream expression pathways, including translation. For example creating or eliminating exons containing upstream open reading frames or altering splicing efficiency and general intron-mediated translation enhancement can dramatically influence the abundance of gene products (20,21). The fraction of gene mutations or variants that influence splicing and gene expression is thus likely to be larger than previously thought. Because copy-number or structural variants exceed single-nucleotides polymorphism by at least 2-fold (22), the overall contribution of human variability to differential pre-mRNA processing and downstream remodelling of ribonucleoprotein particles could be even higher.
Better understanding of the complicated interplay of factors that control splice site choice would clearly benefit from convenient access to a comprehensive resource that pools sequences from scattered mutation reports in a wide-range of biomedical journals. Although rare, these reports provide valuable information about splice site selection in vivo. Currently, the Human Gene Mutation Database (HGMD) (23) and other locus-specific mutation databases (for example HPRT at www.ibiblio.org/dnam/des_hprt.htm or CFTR at www.genet.sickkids.on.ca/cftr/app and see www.hgvs.org/rec.html for other genes) give a list of splicing mutations. However, none of these databases provide critical and comprehensive information needed to understand why aberrant splice sites are selected. Apart from the initial overview of aberrant splicing (24,25), a comprehensive, regularly updated and publicly available tool is missing. Here, we describe DBASS3 and DBASS5, the databases of aberrant 3′-ss and 5′-ss in human disease genes and discuss their utility and importance for studying splice site selection.
RESULTS AND DISCUSSION
Criteria for data inclusion in DBASS3 and DBASS5
Both databases contain sequences of new exon–intron boundaries that were generated through naturally occurring and disease-causing variants or mutations, both germ-line and somatic. DBASS3 and DBASS5 contain mutation-induced and sequence-verified aberrant RNAs published in peer-reviewed communications over the past 30 years (from January 1981 to June 2010). Briefly, reports of cryptic and de novo splice sites were identified by searching PubMed (http://www.ncbi.nlm.nih.gov/entrez/query.fcgi; queries: ‘mutation, splicing, cryptic’; ‘mutation, splicing, new acceptor’ or ‘mutation, splicing, new donor’) and home pages of peer-reviewed journals. A subset of case reports were identified by searching locus-specific mutation databases (http://archive.uwcm.ac.uk/uwcm/mg/docs/oth_mut.html). The search was restricted to human genes with sequence-verified aberrant RNA products.
Reported sequences were manually checked against reference sequences available from human genome databases, including GenBank (http://www.ncbi.nlm.nih.gov/Genbank) (26) and Ensembl (http://www.ensembl.org) (27). The vast majority of aberrant transcripts were verified by amplifying reverse-transcribed total RNA extracted from blood samples taken from affected individuals and/or their family members. Neither database features exon skipping or full intron retention events in which no new exon–intron boundaries were generated, nor do they include polymorphisms that influence utilization of the NAGNAG type of 3′-ss, which was reported elsewhere (28).
Database design and data set summary
DBASS3 and DBASS5 were designed as retrieval and submission tools containing mutation-induced aberrant splice sites that resulted in a recognizable phenotype. The web application was created using the Microsoft ASP and ASP.Net server technology and SQL Server database software. A breakdown of updated DBASS3 and DBASS5 records by gene, phenotype and location of aberrant splice sites is shown in Tables 1 and 2, respectively.
Table 1.
Summary of aberrant 3′-splice sites in DBASS3
| Location of cryptic or de novo 3′-splice sites | Exon |
Intron |
Both | ||
|---|---|---|---|---|---|
| Mutation | In the 3′-ss consensusa (cryptic) | Elsewhere (‘de novo’) | In the 3′-ss consensusa (cryptic) | Elsewhere (‘de novo’) | All mutations |
| Number of genes | 72 | 30 | 39 | 70 | 170 |
| Number of phenotypes | 67 | 32 | 37 | 71 | 165 |
| Number of cryptic and de novo 3′-ss (%) | 107 (34.3) | 48 (15.4) | 49 (15.7) | 108 (34.6) | 312 (100) |
| Number of aberrant 3′-ss affecting terminal exons | 12 | 5 | 9 | 4 | 28 |
| Median distance (nucleotides) between authentic and aberrant 3′-splice sites | 13 | 49 | −41 | −12 | 1 |
aThe 3′-ss consensus is YAG/G (Y is a pyrimidine, slash is the intron–exon boundary).
Table 2.
Summary of aberrant 5′-splice sites in DBASS5
| Location of cryptic or de novo 5′-splice sites | Exon |
Intron |
Both | ||
|---|---|---|---|---|---|
| Mutation | In the 5′-ss consensusa (cryptic) | Elsewhere (‘de novo’) | In the 5′-ss consensusa (cryptic) | Elsewhere (‘de novo’) | All mutations |
| Number of genes | 115 | 57 | 113 | 67 | 255 |
| Number of phenotypes | 122 | 61 | 123 | 68 | 281 |
| Number of cryptic and de novo 5′-ss (%) | 203 (33.9) | 89 (15.2) | 221 (36.9) | 90 (14.0) | 603 (100) |
| Number of aberrant 5′-ss affecting terminal introns (%) | 8 | 2 | 6 | 2 | 18 |
| Median distance (nucleotides) between authentic and aberrant 5′-splice sites | −43 | −59 | 43 | 18 | −9 |
aThe 5′-ss consensus is MAG/GURAGU (M is A or C, R is purine and slash is the exon–intron junction).
The initial analysis of DBASS3 and DBASS5 records (29,30) confirmed that cryptic splice sites were, on average, intrinsically stronger than mutated authentic (natural) sites but generally weaker than their authentic, wild-type counterparts (31). Analysis of DBASS3 and DBASS5 sequences also showed that the maximum entropy (ME) algorithm (32) gave the best overall discrimination between aberrant and authentic sites (29,30), while algorithms based only on the weight matrix, such as the Shapiro and Senapathy score (33) performed less successfully. The density of silencers and enhancers in the segments between authentic and aberrant sites was intermediate between exons and introns, supporting a gradient concept of exon and intron definition (34).
Analysis of DBASS3 and DBASS5 data also revealed the underlying mutation pattern of aberrant splice sites and their location. For example breakdown of DBASS3 alterations showed that AG-creating mutations were much more common than previously estimated, totalling to ∼42% (29), as opposed to the initial estimate of 13% (24). De novo 5′-ss in introns were stronger than their authentic counterparts (30), but this did not apply to exonic de novo sites, suggesting that their activation in vivo is more reliant on exonic splicing enhancers or silencers, rather than on the intrinsic strength of the 5′-ss consensus (30). Finally, DBASS3 and DBASS5 data has been employed to develop other prediction tools, such as CRYP-SKIP (http://www.dbass.org.uk/cryp-skip/), which can distinguish between cryptic splice site activation and exon skipping upon mutation of 3′-ss or 5′-ss (35) or HOT-SKIP (http://www.dbass.org.uk/hot-skip/), which computes the ESS/ESE profile for all possible point mutations at each exon position and identifies nucleotide substitutions that are most likely to skip the exon (M. Raponi et al., submitted for publication). Such free practical utilities are useful for molecular diagnostics to facilitate identification of exonic changes that interfere with RNA processing and prediction of their phenotypic outcome.
User interface
DBASS3 and DBASS5 provide a quick search page where the user needs to specify a gene symbol (http://www.gene.ucl.ac.uk/nomenclature/), phenotype or Mendelian Inheritance in Man (MIM) number (www.ncbi.nlm.nih.gov/omim/) (36) or nucleotide sequences that match the input criteria. In an advanced search option, one can select a combination of several criteria, including phenotype, gene, mutation, location of aberrant splice sites in introns and exons, distance from authentic splice sites and relevant bibliographic references (Figure 1). The user can choose to browse through the list of all records. Investigators studying coupled processes of RNA splicing and end-processing can easily retrieve aberrant splice sites that were activated in terminal exons or introns. Not in the least, the user can search for aberrant splice sites (and their wild-type and mutated authentic counterparts) by their intrinsic strength. This option permits retrieval of splice sites according to various splice-site scores and user-defined cut-off points or desired intervals.
Figure 1.
Screenshot of the DBASS3 search page. DBASS3 and DBASS5 provide a quick search page where the user needs to specify a gene symbol, phenotype or nucleotide sequence. In the Advanced search option, one can select a combination of several criteria, including phenotype, gene, mutation, location of aberrant splice sites in introns and exons, distance from authentic (natural) splice sites, intrinsic strength of splice sites and bibliographic references.
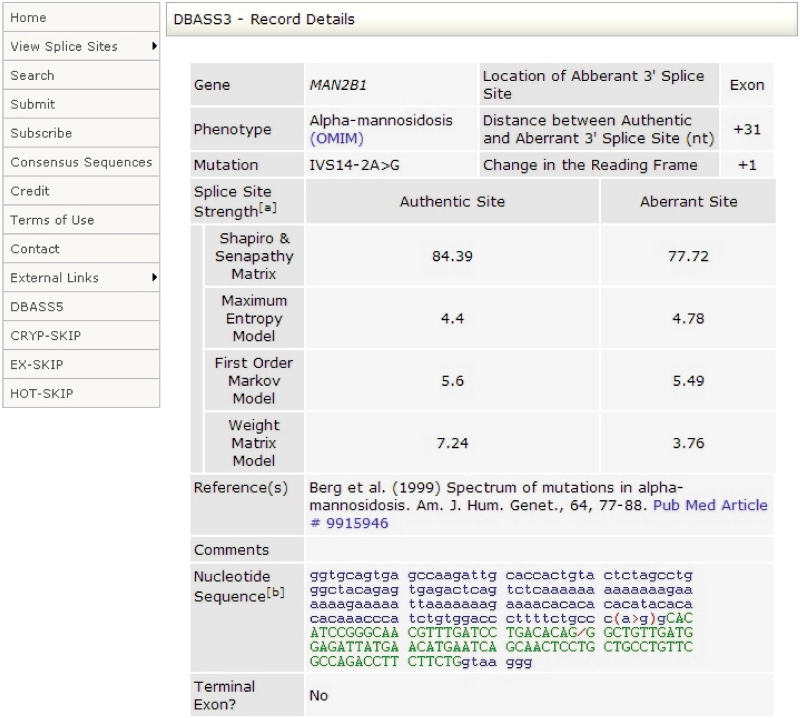
The search results can be expanded by clicking on the ‘View details’ button (Figure 1), leading to full details in each record (Figure 2), including gene, phenotype, MIM numbers, mutation (using traditional and official nomenclature where available), distance between authentic and aberrant splice sites, change of the reading frame (0, +1 and +2 nt), literature references with PubMed hyperlinks and nucleotide sequences flanking the authentic and aberrant 5′-ss. The ex vivo origin of aberrant RNA is indicated in the Comments fields of DBASS3 and DBASS5 records, which also contains additional information.
Figure 2.
Example of a DBASS3 record. Aberrant splice sites are shown as a slash in a genomic sequence. Disease-causing or–—predisposing mutations are denoted by a ‘greater than’ sign for nucleotide substitutions, by parentheses for deletions and by brackets for duplications or insertions. Intronic sequences are shown in blue lower case, exons are shown in green upper case. Cryptic exons are underlined.
Finally, both DBASS3 and DBASS5 allow the users to submit newly published entries for their inclusion in the databases via a dedicated submission tool and also to register to obtain regular database updates by Email.
Nomenclature of aberrant splice sites
Although mutations anywhere in intron or exon can impair pre-mRNA splicing, most mutations that activate aberrant splice sites are found in or close to natural splice sites (29–31). If the mutation nomenclature incorporated the distance (in nucleotides) between mutation and natural exon–intron junction, this information would help to identify mutations that activate aberrant splice sites. For exonic mutations, however, neither traditional (37) nor official (www.hgvs.org/mutnomen/) mutation nomenclature assimilate these data. For intronic alterations, the distance can be derived from both the traditional and the official designation, although the latter does not provide this information when referring to a genomic sequence (for example g.234G>T). As most human mutation reports currently adhere to the official nomenclature, exonic mutations that affect splicing are less likely to be recognized and are probably under-reported. To allow the investigators immediate access to this critical information, both DBASS3 and DBASS5 still show the traditional mutation nomenclature for all records. In exons, single-nucleotide substitutions are simply preceded by ‘E’ (for exon), which is followed by an exon number, a ‘plus’ sign and the distance (in nucleotides) between the last authentic intron–exon junction and the point mutation. In addition to sequence, DBASS3 and DBASS5 users can see both the distance between mutation and authentic splice site and the distance between authentic and aberrant splice sites. Rather than relying merely on the nomenclature, this design ultimately facilitates rapid verification of splicing mutations and auxiliary splicing signals, giving the user immediate access to the sequence of newly intronized or exonized segments and the underlying DNA change.
Future directions
New releases of DBASS3 and DBASS5 data will reflect the growing number of studies reporting new aberrant splice sites. The updates will be facilitated by user-assisted submissions and proper reporting of disease-associated aberrant splices in biomedical journals, which should be endorsed by journal editors. With recent realization that single nucleotide variability is less extensive than insertion/deletion or copy-number variability (22), it will be interesting to catalogue structural alterations that influence pre-mRNA processing and alter the relative expression of pre-existing, alternatively spliced mRNAs, particularly in conserved gene families. DBASS5 and DBASS3 entries are also planned to be linked to external resources other than OMIM and Ensembl, such as the SNP databases (http://www.ncbi.nlm.nih.gov/snp/).
FUNDING
Juvenile Diabetes Research Foundation International (1-2008-47); EC grant EURASNET-LSHG-CT-2005-518238. Funding for open access charge: Juvenile Diabetes Research Foundation, European Union.
Conflict of interest statement. None declared.
REFERENCES
- 1.Sharp PA. Split genes and RNA splicing. Cell. 1994;77:805–815. doi: 10.1016/0092-8674(94)90130-9. [DOI] [PubMed] [Google Scholar]
- 2.Black DL. Protein diversity from alternative splicing: a challenge for bioinformatics and postgenome biology. Cell. 2000;103:367–370. doi: 10.1016/s0092-8674(00)00128-8. [DOI] [PubMed] [Google Scholar]
- 3.Graveley BR. Alternative splicing: increasing diversity in the proteomic world. Trends Genet. 2001;17:100–107. doi: 10.1016/s0168-9525(00)02176-4. [DOI] [PubMed] [Google Scholar]
- 4.Maniatis T, Tasic B. Alternative pre-mRNA splicing and proteome expansion in metazoans. Nature. 2002;418:236–243. doi: 10.1038/418236a. [DOI] [PubMed] [Google Scholar]
- 5.Antonarakis SE, Irkin SH, Cheng TC, Scott AF, Sexton JP, Trusko SP, Charache S, Kazazian HH., Jr beta-Thalassemia in American Blacks: novel mutations in the “TATA” box and an acceptor splice site. Proc. Natl Acad. Sci. USA. 1984;81:1154–1158. doi: 10.1073/pnas.81.4.1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Faustino NA, Cooper TA. Pre-mRNA splicing and human disease. Genes Dev. 2003;17:419–437. doi: 10.1101/gad.1048803. [DOI] [PubMed] [Google Scholar]
- 7.Baralle D, Baralle M. Splicing in action: assessing disease causing sequence changes. J. Med. Genet. 2005;42:737–748. doi: 10.1136/jmg.2004.029538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cooper TA, Wan L, Dreyfuss G. RNA and disease. Cell. 2009;136:777–793. doi: 10.1016/j.cell.2009.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Krawczak M, Thomas NS, Hundrieser B, Mort M, Wittig M, Hampe J, Cooper DN. Single base-pair substitutions in exon-intron junctions of human genes: nature, distribution, and consequences for mRNA splicing. Hum. Mutat. 2007;28:150–158. doi: 10.1002/humu.20400. [DOI] [PubMed] [Google Scholar]
- 10.Mitchell GA, Labuda D, Fontaine G, Saudubray JM, Bonnefont JP, Lyonnet S, Brody LC, Steel G, Obie C, Valle D. Splice-mediated insertion of an Alu sequence inactivates ornithine delta-aminotransferase: a role for Alu elements in human mutation. Proc. Natl Acad. Sci. USA. 1991;88:815–819. doi: 10.1073/pnas.88.3.815. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sorek R. The birth of new exons: mechanisms and evolutionary consequences. RNA. 2007;13:1–6. doi: 10.1261/rna.682507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Vorechovsky I. Transposable elements in disease-associated cryptic exons. Hum. Genet. 2009;127:135–154. doi: 10.1007/s00439-009-0752-4. [DOI] [PubMed] [Google Scholar]
- 13.Yeo G, Hoon S, Venkatesh B, Burge CB. Variation in sequence and organization of splicing regulatory elements in vertebrate genes. Proc. Natl Acad. Sci. USA. 2004;101:15000–15005. doi: 10.1073/pnas.0404901101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Z, Rolish ME, Yeo G, Tung V, Mawson M, Burge CB. Systematic identification and analysis of exonic splicing silencers. Cell. 2004;119:831–845. doi: 10.1016/j.cell.2004.11.010. [DOI] [PubMed] [Google Scholar]
- 15.Zhang XH, Chasin LA. Computational definition of sequence motifs governing constitutive exon splicing. Genes Dev. 2004;18:1241–1250. doi: 10.1101/gad.1195304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Matlin AJ, Clark F, Smith CW. Understanding alternative splicing: towards a cellular code. Nat. Rev. Mol. Cell Biol. 2005;6:386–398. doi: 10.1038/nrm1645. [DOI] [PubMed] [Google Scholar]
- 17.Buratti E, Baralle M, Baralle FE. Defective splicing, disease and therapy: searching for master checkpoints in exon definition. Nucleic Acids Res. 2006;34:3494–3510. doi: 10.1093/nar/gkl498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tasic B, Nabholz CE, Baldwin KK, Kim Y, Rueckert EH, Ribich SA, Cramer P, Wu Q, Axel R, Maniatis T. Promoter choice determines splice site selection in protocadherin alpha and gamma pre-mRNA splicing. Mol. Cell. 2002;10:21–33. doi: 10.1016/s1097-2765(02)00578-6. [DOI] [PubMed] [Google Scholar]
- 19.Furger A, O’Sullivan JM, Binnie A, Lee BA, Proudfoot NJ. Promoter proximal splice sites enhance transcription. Genes Dev. 2002;16:2792–2799. doi: 10.1101/gad.983602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cazzola M, Skoda RC. Translational pathophysiology: a novel molecular mechanism of human disease. Blood. 2000;95:3280–3288. [PubMed] [Google Scholar]
- 21.Kralovicova J, Gaunt TR, Rodriguez S, Wood PJ, Day INM, Vorechovsky I. Variants in the human insulin gene that affect pre-mRNA splicing: is -23HphI a functional single nucleotide polymorphism at IDDM2? Diabetes. 2006;55:260–264. doi: 10.2337/diabetes.55.01.06.db05-0773. [DOI] [PubMed] [Google Scholar]
- 22.Sebat J. Major changes in our DNA lead to major changes in our thinking. Nat. Genet. 2007;39:S3–S5. doi: 10.1038/ng2095. [DOI] [PubMed] [Google Scholar]
- 23.Stenson PD, Mort M, Ball EV, Howells K, Phillips AD, Thomas NS, Cooper DN. The Human Gene Mutation Database: 2008 update. Genome Med. 2009;1:13. doi: 10.1186/gm13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cooper DN, Krawczak M. Human Gene Mutation. Oxford: BIOS Scientific Publishers; 1993. [Google Scholar]
- 25.Nakai K, Sakamoto H. Construction of a novel database containing aberrant splicing mutations of mammalian genes. Gene. 1994;141:171–177. doi: 10.1016/0378-1119(94)90567-3. [DOI] [PubMed] [Google Scholar]
- 26.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Flicek P, Aken BL, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Hiller M, Huse K, Szafranski K, Jahn N, Hampe J, Schreiber S, Backofen R, Platzer M. Single-nucleotide polymorphisms in NAGNAG acceptors are highly predictive for variations of alternative splicing. Am. J. Hum. Genet. 2006;78:291–302. doi: 10.1086/500151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Vorechovsky I. Aberrant 3′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2006;34:4630–4641. doi: 10.1093/nar/gkl535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Buratti E, Chivers MC, Kralovicova J, Romano M, Baralle M, Krainer AR, Vorechovsky I. Aberrant 5′ splice sites in human disease genes: mutation pattern, nucleotide structure and comparison of computational tools that predict their utilization. Nucleic Acids Res. 2007;35:4250–4263. doi: 10.1093/nar/gkm402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Roca X, Sachidanandam R, Krainer AR. Intrinsic differences between authentic and cryptic 5′ splice sites. Nucleic Acids Res. 2003;31:6321–6333. doi: 10.1093/nar/gkg830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Eng L, Coutinho G, Nahas S, Yeo G, Tanouye R, Babaei M, Dork T, Burge C, Gatti RA. Nonclassical splicing mutations in the coding and noncoding regions of the ATM gene: maximum entropy estimates of splice junction strengths. Hum. Mutat. 2004;23:67–76. doi: 10.1002/humu.10295. [DOI] [PubMed] [Google Scholar]
- 33.Shapiro MB, Senapathy P. RNA splice junctions of different classes of eukaryotes: sequence statistics and functional implications in gene expression. Nucleic Acids Res. 1987;15:7155–7174. doi: 10.1093/nar/15.17.7155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kralovicova J, Vorechovsky I. Global control of aberrant splice site activation by auxiliary splicing sequences: evidence for a gradient in exon and intron definition. Nucleic Acids Res. 2007;35:6399–6413. doi: 10.1093/nar/gkm680. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Divina P, Kvitkovicova A, Vorechovsky I. Ab initio prediction of mutation-induced cryptic splice-site activation and exon skipping. Eur. J. Hum. Genet. 2009;17:759–765. doi: 10.1038/ejhg.2008.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Beaudet AL, Tsui LC. A suggested nomenclature for designating mutations. Hum. Mutat. 1993;2:245–248. doi: 10.1002/humu.1380020402. [DOI] [PubMed] [Google Scholar]