Abstract
The HUGO Gene Nomenclature Committee (HGNC) aims to assign a unique gene symbol and name to every human gene. The HGNC database currently contains almost 30 000 approved gene symbols, over 19 000 of which represent protein-coding genes. The public website, www.genenames.org, displays all approved nomenclature within Symbol Reports that contain data curated by HGNC editors and links to related genomic, phenotypic and proteomic information. Here we describe improvements to our resources, including a new Quick Gene Search, a new List Search, an integrated HGNC BioMart and a new Statistics and Downloads facility.
INTRODUCTION
For over thirty years the HUGO Gene Nomenclature committee (HGNC) has striven to aid scientific communication by approving a unique symbol and name for every human gene. The need for a single committee with the authority to approve human gene nomenclature was recognized at the Human Gene Mapping Conference in 1977 and guidelines for naming human genes were subsequently published in 1979 (1). A single dedicated researcher, Prof. Phyllis McAlpine, was initially charged with the enormous task of approving gene symbols, and as the project grew, it was entrusted to a team of post-docs and bioinformaticians under the leadership of Prof. Sue Povey at University College London. Since 2007 the HGNC has been located at the European Bioinformatics Institute (EBI) at Hinxton, Cambridge, UK and our website has been located at www.genenames.org.
The HGNC: our task
The HGNC aims to approve gene symbols and corresponding gene names that are informative, user-friendly and acceptable to researchers in the field. In order to achieve this, we endeavour to contact the researchers that work on particular genes for their advice and input before approving symbols, and encourage researchers to submit proposed gene symbols directly to us to determine their suitability prior to publication. The HGNC team attends conferences regularly to ensure that we are meeting the requirements of the community and to discuss the nomenclature of specific gene families and locus types. We work closely with the nomenclature committees for several other species, especially the mouse (2), rat (3), zebrafish (4) and Xenopus (5) to ensure that orthologous vertebrate genes are assigned equivalent symbols wherever possible. HGNC symbols are used by most biomedical databases, including Ensembl (6), Vega (7), Entrez Gene (8), OMIM (9), GeneCards (10), UCSC (11) and UniProt (12). We maintain a close collaboration with all of these databases: they contact us with information that may be used to approve new gene symbols; we contact them to check the annotation status of genes when necessary.
genenames.org: our resources
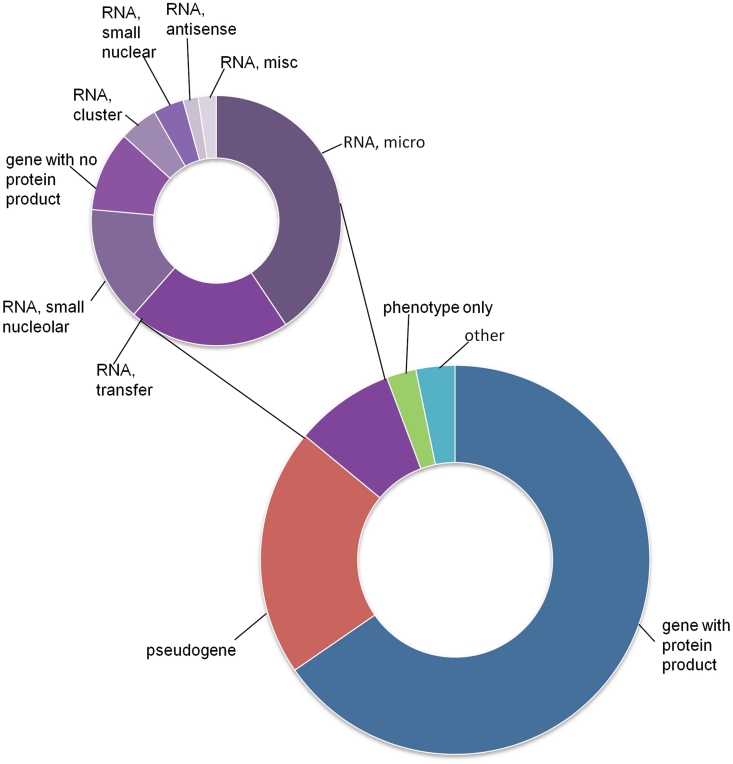
The HGNC website ‘genenames.org’ provides access to all approved human nomenclature and to related genomic, phenotypic and proteomic information, making it a central resource for human genetics. No restrictions are imposed on access to, or use of, the data provided by the HGNC, which are provided to enhance knowledge and encourage progress in the scientific community. As of September 2010, there are almost 30 000 approved gene symbols listed, over 19 000 of which represent protein-coding genes. Each gene with an HGNC-approved symbol has its own Symbol Report that contains our manually-curated core data and links to many other external biomedical resources. The ‘Core Data’ section contains the approved symbol and approved name, and the HGNC ID, a unique number assigned to each gene report that remains stable even if the gene nomenclature is updated. This section also includes previous symbols and names, aliases and the chromosomal location of the gene. Since 2008 (13) we have added the ‘Locus Type’ field to our core data; this field provides information on the genetic class of each gene. The most common locus type is ‘gene with protein product’, which represents 65% of all entries; 19% of entries have the locus type ‘pseudogene’; 8% are classed within the non-coding RNA locus group; 3% are designated as ‘phenotype only’ and the remaining 3% are represented by the locus group ‘other’. This group encompasses locus types that apply to a relatively small number of genes such as ‘immunoglobulin gene’ and ‘T cell receptor gene’ (Figure 1).
Figure 1.
The proportion of HGNC gene symbols annotated with each locus type. The main doughnut chart shows the proportions of major locus groups. The purple region represents genes annotated with the non-protein-coding RNA locus group; the smaller chart shows the proportion of RNA genes annotated with each RNA-specific locus type. The ‘RNA, misc’ category groups together RNA locus types that represent a small number of genes. These are: ‘RNA, Cajal body-specific’, ‘RNA, ribosomal’, ‘RNA, small cytoplasmic’, ‘RNA, vault’, ‘RNA, Y’, ‘RNA, RNase’ and ‘RNA, telomerase’. A full list of locus types, along with total numbers of approved symbols for each category, can be viewed at our Statistics and Downloads facility (www.genenames.org/stats).
The number of genes belonging to the non-protein-coding RNA (ncRNA) locus group has expanded greatly within the last few years. This locus ‘group’ encompasses 13 different RNA locus types such as ‘RNA, antisense’ and ‘RNA, transfer’, making it easy for users to search for and download information on all members of a particular ncRNA subclass. The HGNC is actively engaging the RNA research community in order to provide unique symbols for each ncRNA gene. For instance, we have worked with members of the miRBase project (14) to assign unique symbols for over 1000 pre-miRNA genes and have annotated all of these genes with the locus type ‘RNA, micro’. Another example is our close collaboration with the snoRNABase database (15) which has produced a systematic nomenclature for genes encoding the small nucleolar RNAs (snoRNAs): SNORA# (small nucleolar RNA, H/ACA box containing #) and SNORD# (small nucleolar RNA, C/D box containing #) that are annotated with the locus type ‘RNA, small nucleolar’. The HGNC maintains a dedicated ncRNA gene page on genenames.org, where the complete set of over 2000 ncRNA gene symbols and names can be viewed (www.genenames.org/rna).
As well as the HGNC core data, each Symbol Report contains database IDs and corresponding links to a variety of sequence resources, genome browsers and protein resources, as described earlier (13). Since 2008 we have added Vega IDs with links to the Vega GeneView page (7), CCDS IDs with links to the Consensus CDS project page (16) and RGD IDs with links through to the gene page for the orthologous rat gene at the Rat Genome Database (3) [we have linked via MGI ID to the orthologous gene page at the Mouse Genome Database (2) for many years]. We have also added links through to the relevant webpage of the COSMIC database (17) for genes that are mutated in tumours. Genes that have been implicated in the pathology of rare diseases now link straight through to the Orphanet database (18). We have added two new links that are for genes of particular locus types: pseudogene Symbol Reports contain a link to the annotation page at pseudogene.org (19) where appropriate, and piwi-interacting RNA cluster (PIRC#) Symbol Reports contain a link through to the piRNABank database (20). We have recently added links to searches of the GoPubMed (21) and WikiGenes (22) online databases from all HGNC Symbol reports. The HGNC continues to work closely with Locus-Specific Databases (LSDBs) (23) to ensure that member databases contain approved gene nomenclature. In addition to providing links from over 1300 HGNC Symbol Reports to relevant LSDBs, we have recently created a text file download facility that contains a full list of gene symbols and corresponding LSDB links: see www.genenames.org/lsdb.
In addition to approving gene nomenclature, HGNC editors curate gene family pages; a full list is available at www.genenames.org/genefamily. Genes are grouped into families on the basis of sequence similarity, shared functionality or phenotype. Previously some of these pages were automatically generated based on gene symbol but we have recently updated these so that all our gene family pages are now manually curated. We have over 200 family pages, and over 100 specialist advisors that help us both with the content of the pages and with the approval of new gene family members. Recently, we have organized some pages into superfamilies with subsections for each individual family. For example, the ATPase superfamily page contains the AAA, P-type and Vacuolar-type H+-ATPase (V-ATPase) families, see www.genenames.org/atp.
genenames.org: tools
The genenames.org website contains a number of tools that support searching of HGNC approved nomenclature and related data. We have recently developed a new and improved Quick Gene Search, available from our homepage (www.genenames.org) that provides added functionality compared to the previous simple search. Quick Gene Search accepts multiple keywords (e.g. gene symbols, aliases or parts of gene names) or IDs from the following databases; HGNC, Entrez Gene (8), Ensembl (6), Vega (7), CCDS (16), MGI (2) and RGD (3). There are radio buttons that allow users to search for a result that ‘equals’, ‘contains’ or ‘begins’ with their search term. Quick Gene Search then ranks the results in order of relevance. For example, searching records that contain ‘TP53′ will return the approved gene symbol TP53 at the top of the results list; gene symbols that contain TP53, such as TP53BP1 will rank lower; genes with matching aliases, such as EI24 which has the symbol alias TP53I8, rank further down the results list; and genes with TP53 in the name such as ‘PERP, TP53 apoptosis effector’ rank further still down the list. Quick Gene Search results are now also paginated so that users can access all results easily. Our Advanced Search (www.genenames.org/advancedsearch) is being updated with extra functionalities which will also include the ranking and pagination of results.
List search
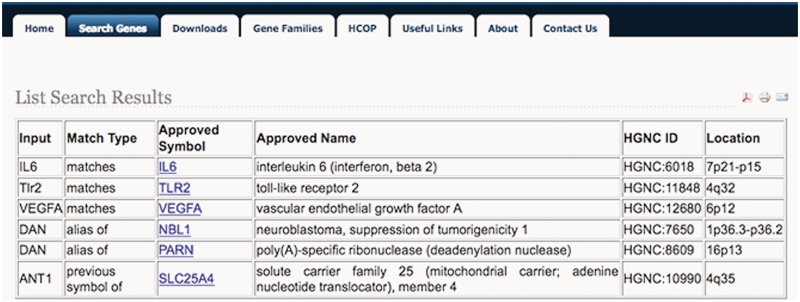
We have also recently developed the HGNC List Search (www.genenames.org/list) which allows searching of multiple gene symbols in one step. Lists of symbols can be typed, pasted or uploaded directly into the tool. Figure 2 shows an example of the List Search results output. The results include a ‘match type’ column that shows how each submitted symbol matches the returned HGNC symbol. For example, the search term IL6 ‘matches’ the approved symbol IL6, and ANT1 matches as a ‘previous symbol of’ the approved symbol SLC25A4. The basic version of the tool is case insensitive so the search term Tlr2 ‘matches’ the approved symbol TLR2. The search term DAN is an ‘alias of’ both the approved symbol NBL1 and the approved symbol PARN, so two sets of results are returned for this term; the user is able to click on the approved symbol to be taken to the relevant Symbol Report to access more information on the two possible gene symbols. An advanced version of this tool is also available (www.genenames.org/bulkcheck) that supports case sensitive searching and allows results to be downloaded as text.
Figure 2.
An example of the results output generated by the HGNC List Search. The ‘Match Type’ column shows how each submitted symbol is related to the matched gene entry. Each approved symbol is hyperlinked to the HGNC Symbol Report so that users can view further information on the matched genes.
HCOP
The HGNC Comparison of Orthology Predictions (HCOP) tool (www.genenames.org/hcop) aggregates orthology predictions between human and 14 different species from a range of data sources (24). Therefore, HCOP provides a single resource for comparison of orthology data, enabling users to identify consensus orthology predictions quickly from the displayed data. Since 2008 (13) HCOP has been updated with orthology calls between human and cow, Caenorhabditis elegans, Saccharomyces cerevisiae, platypus, macaque, opossum and horse, and with source data from UCSC (11) and the OPTIC (Orthologous and Paralogous Transcripts in Clades) database (25). HCOP can be searched for a specified gene, or set of genes, using approved symbols, Entrez Gene IDs, HGNC IDs, MGI IDs or RefSeq accessions. In addition to the orthology predictions, HCOP results contain a link back to the source database for each assertion; our source databases are Ensembl (6), Evola (26), HGNC, HomoloGene (27), Inparanoid (28), MGI (2), PhyOP (29), Treefam (30), OPTIC (25) and UCSC (11). There is also a link to the Entrez Gene (8) page for each listed ortholog. The results for orthologs from species with a gene nomenclature committee [mouse (2), rat (3), chicken (31), zebrafish (4), Drosophila (32), C. elegans (33) and S. cerevisiae (34)] display the approved symbol and a link to the appropriate nomenclature database. For other species currently without an official naming authority (chimp, macaque, dog, horse, cow and platypus) the displayed gene symbols are derived from Entrez Gene (8). We have recently updated the tool with a new text mode output to return results as a tab delimited file. Additionally, HCOP contains a Bulk Downloads section that provides the complete orthology assertion data for each species set as text files.
Statistics and downloads
The HGNC Statistics and Downloads facility (www.genenames.org/stats) provides access to the full HGNC data set and to specific subdivisions of data either by broad locus group e.g. ‘non-protein-coding RNA’ or by specific locus type e.g. ‘RNA, small nuclear’. The page also includes statistics on the total number of approved symbols per data set. There is a quick link to a tab delimited text file containing the core data for each data subdivision. Each data set also has a link to the Custom Downloads page, a web-based interface that allows users to select exactly which data fields to download and to choose between output formats including tab delimited text file and html table. The Custom Downloads tool can also be used to generate Perl code to automate downloading subsets of specified HGNC data.
BioMart and EB-eye
In 2008, the HGNC launched a BioMart tool (www.genenames.org/biomart). This provides an alternative open source means of accessing HGNC data via the BioMart web interface, Perl API, RESTful web service, SOAP web service and a DAS server (35). The tool allows users to perform complex queries and to choose exactly which data fields are included in the results. Results are returned as HTML, comma separated values (CSV) or tab separated values (TSV). The BioMart interface at genenames.org queries HGNC data only but the MartView at the BioMart Central Portal (www.biomart.org) supports queries that combine the HGNC data set with other data sets such as Ensembl (6), Vega (7), MGI (2) and RGD (3). HGNC data has also recently been integrated into the EB-eye search tool (www.ebi.ac.uk/ebisearch), a one-step search engine for all biological data held at the EBI (36). HGNC results can be found in the ‘Genomes’ section of the EB-eye results table.
genenames.org: future directions
We are currently redesigning our website to make navigation more intuitive. Each page on genenames.org will include a tabbed navigation menu with dropdown menus to access all of our tools and pages, as well as a site-wide text search and links to submit gene symbol requests and feedback. On the updated homepage the new Quick Gene Search will feature prominently, along with updated FAQs and a ‘News’ section. We are also reformatting our Symbol Report pages to be consistent in design with our new website.
In the future we will expand our gene family resources to include more families and groupings, further links to external databases, and information on the predicted protein architecture. We will focus on approving nomenclature for pseudogenes, the majority of which remain largely unnamed, and continue to provide approved symbols for non-coding RNAs, especially for the currently under-represented long (>200 nt) non-coding RNAs. We also look forward to working with other database, nomenclature and genome groups to support the assignment of consistent nomenclature for orthologs across all vertebrate species (37). As part of this initiative, we aim to reassign human genes that have anonymous C#orf# (chromosome # open reading frame #) symbols with new symbols based on function and sequence characteristics where possible. To be notified of all upcoming changes and updates to our project please subscribe to our newsletter by contacting hgnc@genenames.org using the subject line ‘subscribe’ and including your Email address.
FUNDING
The Wellcome Trust (081979/Z/07/Z); National Human Genome Research Institute (P41 HG03345). Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Louise Daugherty for her helpful comments on the content of this article.
REFERENCES
- 1.Shows TB, Alper CA, Bootsma D, Dorf M, Douglas T, Huisman T, Kit S, Klinger HP, Kozak C, Lalley PA, et al. International system for human gene nomenclature (1979) ISGN (1979) Cytogenet. Cell Genet. 1979;25:96–116. doi: 10.1159/000131404. [DOI] [PubMed] [Google Scholar]
- 2.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. The Rat Genome Database, update 2007–easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sprague J, Bayraktaroglu L, Bradford Y, Conlin T, Dunn N, Fashena D, Frazer K, Haendel M, Howe DG, Knight J, et al. The Zebrafish Information Network: the zebrafish model organism database provides expanded support for genotypes and phenotypes. Nucleic Acids Res. 2008;36:D768–D772. doi: 10.1093/nar/gkm956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bowes JB, Snyder KA, Segerdell E, Jarabek CJ, Azam K, Zorn AM, Vize PD. Xenbase: gene expression and improved integration. Nucleic Acids Res. 2010;38:D607–D612. doi: 10.1093/nar/gkp953. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wilming LG, Gilbert JG, Howe K, Trevanion S, Hubbard T, Harrow JL. The vertebrate genome annotation (Vega) database. Nucleic Acids Res. 2008;36:D753–D760. doi: 10.1093/nar/gkm987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Maglott D, Ostell J, Pruitt KD, Tatusova T. Entrez Gene: gene-centered information at NCBI. Nucleic Acids Res. 2007;35:D26–D31. doi: 10.1093/nar/gkl993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37:D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Safran M, Dalah I, Alexander J, Rosen N, Iny Stein T, Shmoish M, Nativ N, Bahir I, Doniger T, Krug H, et al. GeneCards Version 3: the human gene integrator. Database. 2010;2010 doi: 10.1093/database/baq020. doi:10.1093 [Epub ahead of print, 7 August 2010] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36:D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Griffiths-Jones S. miRBase: microRNA sequences and annotation. Curr. Protoc. Bioinformatics. 2010 doi: 10.1002/0471250953.bi1209s29. Chapter 12, Unit 12 19 11–10. [DOI] [PubMed] [Google Scholar]
- 15.Lestrade L, Weber MJ. snoRNA-LBME-db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res. 2006;34:D158–D162. doi: 10.1093/nar/gkj002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Weinreich SS, Mangon R, Sikkens JJ, Teeuw ME, Cornel MC. [Orphanet: a European database for rare diseases] Ned. Tijdschr. Geneeskd. 2008;152:518–519. [PubMed] [Google Scholar]
- 19.Karro JE, Yan Y, Zheng D, Zhang Z, Carriero N, Cayting P, Harrrison P, Gerstein M. Pseudogene.org: a comprehensive database and comparison platform for pseudogene annotation. Nucleic Acids Res. 2007;35:D55–D60. doi: 10.1093/nar/gkl851. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Sai Lakshmi S, Agrawal S. piRNABank: a web resource on classified and clustered Piwi-interacting RNAs. Nucleic Acids Res. 2008;36:D173–D177. doi: 10.1093/nar/gkm696. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Doms A, Schroeder M. GoPubMed: exploring PubMed with the Gene Ontology. Nucleic Acids Res. 2005;33:W783–W786. doi: 10.1093/nar/gki470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hoffmann R. A wiki for the life sciences where authorship matters. Nat. Genet. 2008;40:1047–1051. doi: 10.1038/ng.f.217. [DOI] [PubMed] [Google Scholar]
- 23.Horaitis O, Talbot CC, Jr, Phommarinh M, Phillips KM, Cotton RG. A database of locus-specific databases. Nat. Genet. 2007;39:425. doi: 10.1038/ng0407-425. [DOI] [PubMed] [Google Scholar]
- 24.Eyre TA, Wright MW, Lush MJ, Bruford EA. HCOP: a searchable database of human orthology predictions. Brief Bioinform. 2007;8:2–5. doi: 10.1093/bib/bbl030. [DOI] [PubMed] [Google Scholar]
- 25.Heger A, Ponting CP. OPTIC: orthologous and paralogous transcripts in clades. Nucleic Acids Res. 2008;36:D267–D270. doi: 10.1093/nar/gkm852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Matsuya A, Sakate R, Kawahara Y, Koyanagi KO, Sato Y, Fujii Y, Yamasaki C, Habara T, Nakaoka H, Todokoro F, et al. Evola: ortholog database of all human genes in H-InvDB with manual curation of phylogenetic trees. Nucleic Acids Res. 2008;36:D787–D792. doi: 10.1093/nar/gkm878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Geer LY, Marchler-Bauer A, Geer RC, Han L, He J, He S, Liu C, Shi W, Bryant SH. The NCBI BioSystems database. Nucleic Acids Res. 2010;38:D492–D496. doi: 10.1093/nar/gkp858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ostlund G, Schmitt T, Forslund K, Kostler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Goodstadt L, Ponting CP. Phylogenetic reconstruction of orthology, paralogy, and conserved synteny for dog and human. PLoS Comput. Biol. 2006;2:e133. doi: 10.1371/journal.pcbi.0020133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ruan J, Li H, Chen Z, Coghlan A, Coin LJ, Guo Y, Heriche JK, Hu Y, Kristiansen K, Li R, et al. TreeFam: 2008 Update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Burt DW, Carre W, Fell M, Law AS, Antin PB, Maglott DR, Weber JA, Schmidt CJ, Burgess SC, McCarthy FM. The Chicken Gene Nomenclature Committee report. BMC Genomics. 2009;10(Suppl. 2):S5. doi: 10.1186/1471-2164-10-S2-S5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Harris TW, Antoshechkin I, Bieri T, Blasiar D, Chan J, Chen WJ, De La Cruz N, Davis P, Duesbury M, Fang R, et al. WormBase: a comprehensive resource for nematode research. Nucleic Acids Res. 2010;38:D463–D467. doi: 10.1093/nar/gkp952. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Engel SR, Balakrishnan R, Binkley G, Christie KR, Costanzo MC, Dwight SS, Fisk DG, Hirschman JE, Hitz BC, Hong EL, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38:D433–D436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal–unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Valentin F, Squizzato S, Goujon M, McWilliam H, Paern J, Lopez R. Fast and efficient searching of biological data resources–using EB-eye. Brief Bioinform. 2010;11:375–384. doi: 10.1093/bib/bbp065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bruford EA. Highlights of the ‘gene nomenclature across species' meeting. Hum. Genomics. 2010;4:213–217. doi: 10.1186/1479-7364-4-3-213. [DOI] [PMC free article] [PubMed] [Google Scholar]