Abstract
Despite the availability of a large number of protein–protein interactions (PPIs) in several species, researchers are often limited to using very small subsets in a few organisms due to the high prevalence of spurious interactions. In spite of the importance of quality assessment of experimentally determined PPIs, a surprisingly small number of databases provide interactions with scores and confidence levels. We introduce HitPredict (http://hintdb.hgc.jp/htp/), a database with quality assessed PPIs in nine species. HitPredict assigns a confidence level to interactions based on a reliability score that is computed using evidence from sequence, structure and functional annotations of the interacting proteins. HitPredict was first released in 2005 and is updated annually. The current release contains 36 930 proteins with 176 983 non-redundant, physical interactions, of which 116 198 (66%) are predicted to be of high confidence.
INTRODUCTION
Protein–protein interactions (PPIs) are vital for cellular function in organisms and hence their detection is of considerable importance. The advent of high-throughput technologies has lead to a manifold increase in the PPI information in several model organisms through large scale yeast two hybrid (Y2H) and tandem affinity purification in combination with mass spectrometry (TAP/MS) experiments. However, this data has two major drawbacks leading to its limited usage—(i) the large number of spurious interactions detected (1) and (ii) the absence of direct binary interaction information in protein co-complex data obtained from TAP/MS experiments (2). As a result, most studies using PPI information either use data obtained exclusively from small-scale experiments, or those confirmed in multiple experiments. Both types of interaction subsets are considered high confidence but constitute only a fraction of the amount of data available (3) and their use can often lead to biased results. An alternative approach is to utilize the high confidence subsets provided by authors of high-throughput experiments. However, these interaction subsets are assessed using a range of techniques with differing accuracies making comparisons among data sets difficult. Frequently, such high confidence interaction subsets are available only for one or two species, typically yeast and human. As a result, a large amount of the PPI information in several species, though correct and potentially useful, is often ignored.
The major reason for this lack of information usage is the scarcity of comprehensive PPI databases that provide confidence scores assessing the quality of the interactions. Of the many PPI databases that are currently in use [IntAct (4), BioGRID (5), BIND (6), MINT (7), DIP (8), STRING (9), MPPI (10), HPRD (11), MPACT (12) and consolidated databases like iRefWeb (13) and APHID (14)], only two provide confidence scores, namely STRING and MINT. The score in MINT relies on the number and types of experiments in which the interaction is detected without adequately utilizing the genomic annotations of the interacting proteins. STRING uses genomic association information along with homology, annotation and experiment information, but does not consider information regarding interacting domains. Furthermore, in spite of the development of a number of methods to assess interaction quality, there is no consensus on the best method and few are actually applied to multiple large interaction data sets in more than one species, or make the high confidence data sets easily accessible (15–20).
To address these issues, we introduce HitPredict (http://hintdb.hgc.jp/htp/), a database of quality assessed interactions in nine species. HitPredict combines interactions from IntAct, BioGRID and HPRD and determines the confidence level of the interactions based on a reliability score calculated using the sequence, structure and functional annotations of the interacting proteins (21). HitPredict was first introduced in 2005 as a database of high confidence PPIs from high-throughput data sets. It has since been updated annually and has now been expanded to include small-scale interactions along with a more intuitive user interface.
DATABASE CONTENT
HitPredict contains 176 983 non-redundant, physical PPIs among 36 930 proteins, collated from IntAct, BioGRID and the HPRD. We selected these three databases because they have high data coverage and comprehensive annotations. Genetic interactions and those among proteins with obsolete identifiers in UniProt (22) were excluded. Annotations and links to external databases are extensively provided. Coexpression correlation coefficients of interacting proteins obtained from COXPRESdb (23) and ATTED-II (24) are also assigned for mammals and plants, respectively.
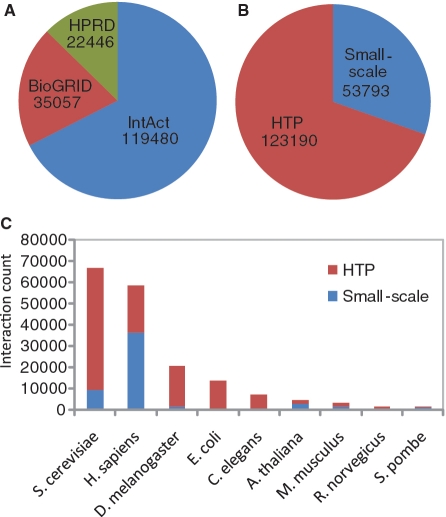
Interactions in HitPredict are differentiated into two types—small-scale and high-throughput, depending on the nature of the experiment in which they were identified. The distinction between small-scale and high-throughput experiments is ambiguous but critical, primarily because interactions from small-scale experiments are typically considered to be of high confidence. For the purposes of HitPredict, experiments with <100 interactions are considered to be small-scale and high confidence, while the rest are denoted as high-throughput. This cutoff value is based on the observation that ∼90% of the interactions in experiments with <100 interactions are supported by multiple evidences (See Supplementary Data for details). The interactions are further categorized into directly observed binary interactions and those derived from protein co-complex data using the spoke model (i.e. bait interacts with each of the prey proteins). Figure 1 shows the distribution of interactions in HitPredict by source, type and species. The large number of high-throughput interactions emphasizes the need for quality assessment.
Figure 1.
Distribution of experimentally determined and quality assessed PPIs in HitPredict (A) by source database, (B) by experiment type—small-scale or high-throughput (HTP), (C) by species and experiment type.
QUALITY ASSESSMENT
All high-throughput interactions and small-scale interactions derived from co-complex data are assessed for their reliability. Interactions from small-scale binary experiments are considered to be high confidence without the assignment of a score (See Supplementary Data for benchmark). As described in detail in our previous report (21), HitPredict calculates the reliability of interactions in the form of a likelihood ratio using naïve Bayesian networks to combine evidence from the presence of the following features:
interacting proteins contain Pfam domains known to interact in complex structures in Protein Data Bank, as obtained from the 3DID database, which uses an empirical scoring scheme to filter out artifacts from crystal packing (25);
interacting proteins share at least one common Gene Ontology term (26); and
the interaction has homologous interactions in the same or other species, as given by the Hintdb database, which identifies homologs using PSIBlast with five iterations and an e-value cutoff of 10−8 (27).
An evaluation of the quality of prediction of the features shows that interacting Pfam domains is the most accurate, followed by common GO terms and homologous interactions respectively. The combined likelihood ratio from these features is an estimate of the posterior odds of an interaction, with one or more features, being true. A likelihood ratio greater than 1 indicates that the interaction is supported by one or more of the features and thus has a greater probability of being true. This method has good specificity and sensitivity in the confidence predictions made (21). Additionally, this scoring scheme differs from that in STRING or MINT since it does not depend on the number of experiments supporting an interaction or the number of interactions determined in a data set, and uses domain–domain interaction information with other genomic features. This makes HitPredict especially useful in identifying high confidence subsets in interactions detected in a single high-throughput experiment. Thus, it potentially provides an alternative perspective on the quality of the interaction data. This is confirmed by comparison of the total and high confidence interactions in Saccharomyces cerevisiae in HitPredict, STRING and MINT (See Supplementary Data).
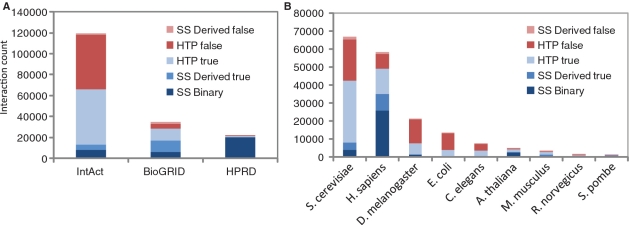
Of the 176 983 PPIs in HitPredict, 116 198 (66%) are predicted to be of high confidence. The breakup of predicted error rates in PPIs obtained from different data sources and in different species is shown in Figure 2 and Supplementary Table S1. The presence of a large number of low confidence interactions in several data sets highlights the need for databases like HitPredict. Supplementary Figure S1 gives the percentage of high confidence interactions in HitPredict for 23 high-throughput experiments with more than 1000 interactions, published from 2000 to 2009, and shows the large number of predicted false positives in many of them. The large number of high confidence interactions predicted in high quality data sets like Collins et al. (28) illustrates the good performance of HitPredict.
Figure 2.
PPI data accuracy as determined by HitPredict in data obtained (A) from different sources, where the interaction accuracy does not reflect the accuracy of the database itself, but of the data in HitPredict that is obtained from the database, (B) in different species. False positive or low confidence interactions are shown in shades of red, whereas high confidence interactions are displayed in shades of blue. Shades denote the experiment type. SS- small-scale; HTP- high-throughput; Binary- interaction obtained from Y2H or other direct detection method; Derived- interaction obtained from protein co-complex data expanded using the spoke model.
USAGE
HitPredict can be used for three main purposes.
Determining the high confidence interactions of a protein
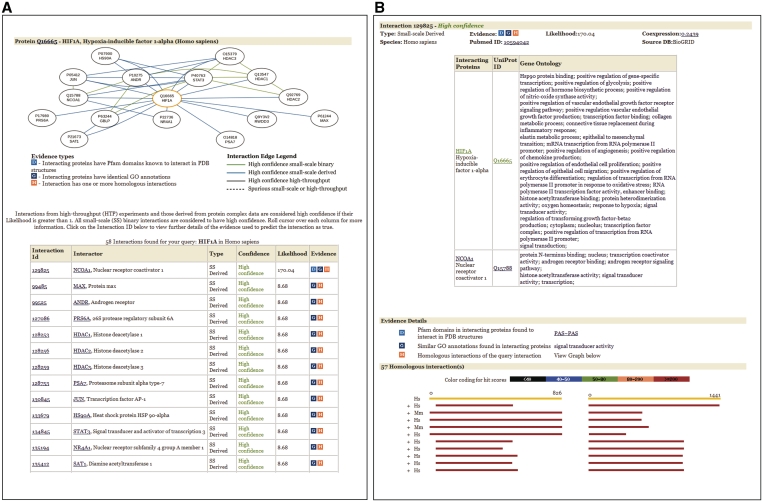
Interactions for proteins can be searched for using a number of protein identifiers like UniProt ID, Entrez Gene ID, RefSeq Protein ID, the protein name or a description keyword. Selecting the protein from the results displays the interactions of the protein as a graphical network and a table (Figure 3A). The graph shows the interaction network of the query protein and its interacting partners. The color and style of the link indicates the quality of the interaction and the type of experiment in which it was detected. The table of interaction partners contains details of the confidence assigned to each interaction, the score in the form of the likelihood ratio, and the supporting evidence used to determine the score (Figure 3A). Details of individual interactions and the evidence supporting them can be seen by selecting the interaction of interest. This leads to a page giving details and annotations for the interaction, such as the source database and publications, the co-expression correlation coefficient of the genes and the protein annotations (Firgure 3B). The evidence details shown include the Pfam domains in the interacting proteins which are known to interact in 3D structures, the common GO terms and a graphical display of the homologous interactions showing the species, the score, e-value and percent identity of the homologous proteins.
Figure 3.
(A) Interactions for a protein in HitPredict. Selecting a protein from the search results bring the user to this page. The graphical view shows the protein as an orange node and its interaction partners in black. Clicking on a node leads to the interactions page of that protein. Interactions are indicated by the links in the graph and color coded by type as given in the Legend. The table underneath the graph gives details of each interaction, the type of experiment it was detected in, the predicted confidence level, the score in the form of the Likelihood and the evidence. Evidence is given as D—structurally known interacting domains, G—common GO terms in interacting proteins and H—presence of homologous interactions. (B) Interaction details and evidence supporting the quality assessment. The interaction annotations, confidence and evidence are given here. Evidence details in the form of interacting Pfam domains, common GO terms and homologous interactions are shown.
For example, in order to find the high confidence interactions of the protein ‘HIF1A’, the hypoxia-inducible factor 1-α, in humans, searching for the term ‘hif1’ produces proteins in several species. Selecting the protein ‘hif1a_human’ from the search results leads a page showing 58 interactions for hif1a_human, of which, 52 are of high confidence. The interactions obtained from HitPredict can be compared to those in STRING (66 interactions of which 15 are high confidence interactions with a score >0.7) and MINT (26 interactions of which 16 may be considered high confidence with a score >0.4). In this case, the high confidence data set provided by HitPredict contains interactions over and above those from MINT and STRING. However, this may not always be the case since the number of interactions and the scoring scheme vary among databases (Supplementary Data). Thus, referring to multiple databases with distinct scoring schemes is a prudent approach.
Identifying the high-confidence interactions from a specific experiment
Interactions of experiments in HitPredict, specifically high-throughput ones, can be directly searched for using the Pubmed ID. A list of these is provided in the Help section for user reference. The resulting interactions are displayed in a tabular form (Figure 3A), and interaction and evidence details can be viewed as described in previous section (Figure 3B). This feature is currently not available in STRING.
Obtaining large high confidence data sets for computational analyses
High confidence interactions from small-scale experiments can be downloaded and used either for network analysis or as gold standard data sets. Other predicted high confidence interactions can be downloaded and used to analyze large interaction networks or specific sub-networks in combination with additional data such as for transcription factors or disease genes. High confidence interaction data downloaded from HitPredict is easy to use since it is categorized by species and type of the interactions and includes the interaction details, the score and evidence used to compute the score. The use of UniProt identifiers makes it convenient to map the interacting proteins onto other protein identifiers, and annotate them. This makes it easier to use than the data downloaded from MINT, which does not include the confidence scores for all interactions, or STRING, which does not include the Pubmed IDs and uses different types of protein identifiers in different species.
Thus, HitPredict can be used to confirm the interactions of a small set of proteins or to perform large-scale feature analyses of high confidence interaction networks. It may either be used independently or in combination with other databases that provide confidence scores.
DISCUSSION
The use of multiple genomic features to calculate a reliability score, the availability of high confidence interaction subsets in several species, and the ease of obtaining these scored interaction subsets, are some of the advantages of HitPredict. It provides an additional means of identifying high confidence PPIs using an alternative scoring strategy. The use of a common scoring scheme for interactions from different experiments allows the comparison of multiple data sets and sources.
Specifically, as compared to MINT, HitPredict provides a larger interaction set, quality assessment scores for all high-throughput interactions and use of multiple genomic features for score calculation. In comparison to STRING, HitPredict provides the ability to search interactions from an experiment using Pubmed ID, uniform annotations using UniProt identifiers for easier mapping across databases, and categorized interaction files facilitating data download and large-scale analyses. Additionally, it uses information from structurally known interacting Pfam domains in the quality assessment. The presence of non-specific interactions through crystal contacts in 3DID, and the possibility that in some cases the proteins with these domains may interact differently than previously observed, seems to have a minimal effect on the performance of this feature. Indeed, this feature has the highest reliability in predicting high quality interactions indicating the minimal effect of non-specific domain interactions and confirming the previous finding that homologs of interacting protein pairs interact in a similar manner (29).
Future enhancements include incorporation of data from additional PPI databases, further annotations for proteins and interactions, and improvement of the confidence score by including experiment number and type information. User interface enhancements will enable users to view larger interaction networks in graphical format, and display homologs of proteins with links to their interactions. HitPredict updates are currently performed once a year. HitPredict has been continually maintained, updated and enhanced in the last 5 years in order to make it a comprehensive and easily accessible source of quality-assessed PPIs in multiple species.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: Japan Society for the Promotion of Science (JSPS) through its Funding Program for World-Leading Innovative R&D in Science and Technology (FIRST Program).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Riu Yamashita (University of Tokyo) for help with running PSIBlast on the Super Computer System, and Dr Takeshi Obayashi (Tohoku University) for help with preparing the interaction network graphs. Computation time is provided by the Super Computer System, Human Genome Center, Institute of Medical Science, University of Tokyo.
REFERENCES
- 1.Bork P, Jensen LJ, von Mering C, Ramani AK, Lee I, Marcotte EM. Protein interaction networks from yeast to human. Curr. Opin. Struct. Biol. 2004;14:292–299. doi: 10.1016/j.sbi.2004.05.003. [DOI] [PubMed] [Google Scholar]
- 2.Wodak SJ, Pu S, Vlasblom J, Seraphin B. Challenges and rewards of interaction proteomics. Mol. Cell. Proteomics. 2009;8:3–18. doi: 10.1074/mcp.R800014-MCP200. [DOI] [PubMed] [Google Scholar]
- 3.von Mering C, Krause R, Snel B, Cornell M, Oliver SG, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 4.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Breitkreutz B-J, Stark C, Reguly T, Boucher L, Breitkreutz A, Livstone M, Oughtred R, Lackner DH, Bahler J, Wood V, et al. The BioGRID interaction database: 2008 update. Nucleic Acids Res. 2008;36:D637–D640. doi: 10.1093/nar/gkm1001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Bader GD, Betel D, Hogue CW. BIND: the biomolecular interaction network database. Nucleic Acids Res. 2003;31:248–250. doi: 10.1093/nar/gkg056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ceol A, Chatr Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38:D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Pagel P, Kovac S, Oesterheld M, Brauner B, Dunger-Kaltenbach I, Frishman G, Montrone C, Mark P, Stumpflen V, Mewes H-W, et al. The MIPS mammalian protein-protein interaction database. Bioinformatics. 2005;21:832–834. doi: 10.1093/bioinformatics/bti115. [DOI] [PubMed] [Google Scholar]
- 11.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Guldener U, Munsterkotter M, Oesterheld M, Pagel P, Ruepp A, Mewes H-W, Stumpflen V. MPact: the MIPS protein interaction resource on yeast. Nucleic Acids Res. 2006;34:D436–D441. doi: 10.1093/nar/gkj003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Razick S, Magklaras G, Donaldson I. iRefIndex: a consolidated protein interaction database with provenance. BMC Bioinformatics. 2008;9:405. doi: 10.1186/1471-2105-9-405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Prieto C, De Las Rivas J. APID: agile protein interaction dataanalyzer. Nucleic Acids Res. 2006;34:W298–W302. doi: 10.1093/nar/gkl128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Deane CM, Salwinski L, Xenarios I, Eisenberg D. Protein interactions: two methods for assessment of the reliability of high throughput observations. Mol. Cell Proteomics. 2002;1:349–356. doi: 10.1074/mcp.m100037-mcp200. [DOI] [PubMed] [Google Scholar]
- 16.Sprinzak E, Sattath S, Margalit H. How reliable are experimental protein-protein interaction data? J. Mol. Biol. 2003;327:919–923. doi: 10.1016/s0022-2836(03)00239-0. [DOI] [PubMed] [Google Scholar]
- 17.Saito R, Suzuki H, Hayashizaki Y. Construction of reliable protein-protein interaction networks with a new interaction generality measure. Bioinformatics. 2003;19:756–763. doi: 10.1093/bioinformatics/btg070. [DOI] [PubMed] [Google Scholar]
- 18.Bader JS, Chaudhuri A, Rothberg JM, Chant J. Gaining confidence in high-throughput protein interaction networks. Nat. Biotech. 2004;22:78–85. doi: 10.1038/nbt924. [DOI] [PubMed] [Google Scholar]
- 19.Li D, Liu W, Liu Z, Wang J, Liu Q, Zhu Y, He F. PRINCESS, a protein interaction confidence evaluation system with multiple data sources. Mol. Cell Proteomics. 2008;7:1043–1052. doi: 10.1074/mcp.M700287-MCP200. [DOI] [PubMed] [Google Scholar]
- 20.Kiemer L, Costa S, Ueffing M, Cesareni G. WI-PHI: a weighted yeast interactome enriched for direct physical interactions. Proteomics. 2007;7:932–943. doi: 10.1002/pmic.200600448. [DOI] [PubMed] [Google Scholar]
- 21.Patil A, Nakamura H. Filtering high-throughput protein-protein interaction data using a combination of genomic features. BMC Bioinformatics. 2005;6:100. doi: 10.1186/1471-2105-6-100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.The UniProt Consortium. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Obayashi T, Hayashi S, Shibaoka M, Saeki M, Ohta H, Kinoshita K. COXPRESdb: a database of coexpressed gene networks in mammals. Nucleic Acids Res. 2008;36:D77–D82. doi: 10.1093/nar/gkm840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Obayashi T, Hayashi S, Saeki M, Ohta H, Kinoshita K. ATTED-II provides coexpressed gene networks for Arabidopsis. Nucleic Acids Res. 2009;37:D987–D991. doi: 10.1093/nar/gkn807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stein A, Panjkovich A, Aloy P. 3did Update: domain-domain and peptide-mediated interactions of known 3D structure. Nucleic Acids Res. 2009;37:D300–D304. doi: 10.1093/nar/gkn690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Patil A, Nakamura H. HINT - a database of annotated protein-protein interactions and their homologs. BIOPHYSICS. 2005;1:21–24. doi: 10.2142/biophysics.1.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Collins SR, Kemmeren P, Zhao XC, Greenblatt JF, Spencer F, Holstege FC, Weissman JS, Krogan NJ. Toward a comprehensive atlas of the physical interactome of Saccharomyces cerevisiae. Mol. Cell Proteomics. 2007;6:439–450. doi: 10.1074/mcp.M600381-MCP200. [DOI] [PubMed] [Google Scholar]
- 29.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc. Natl Acad. Sci. USA. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]