Abstract
The molecular diversity of viruses complicates the interpretation of viral genomic and proteomic data. To make sense of viral gene functions, investigators must be familiar with the virus host range, replication cycle and virion structure. Our aim is to provide a comprehensive resource bridging together textbook knowledge with genomic and proteomic sequences. ViralZone web resource (www.expasy.org/viralzone/) provides fact sheets on all known virus families/genera with easy access to sequence data. A selection of reference strains (RefStrain) provides annotated standards to circumvent the exponential increase of virus sequences. Moreover ViralZone offers a complete set of detailed and accurate virion pictures.
INTRODUCTION
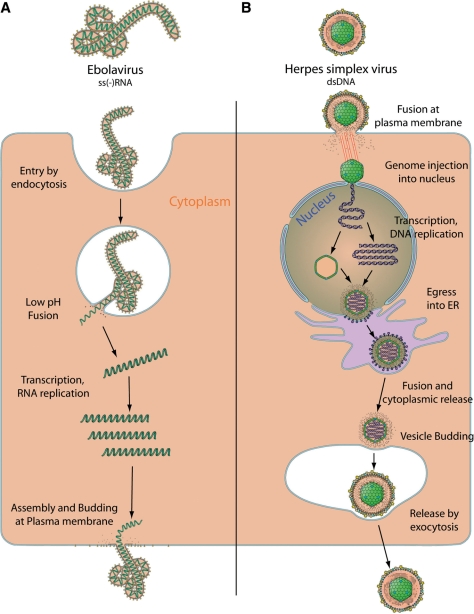
Viruses are presumably the most abundant biological entities on the planet, with the total number of virus particles exceeding by 10 times the total number of cells (25). Many viruses have a relatively small genome encoding for a few proteins: one of the smallest being the circovirus with a 1.7-kb genome coding only two proteins (11). Despite their apparent simplicity, viral biochemistry and replication mechanisms are more varied than those seen in the entire bacterial, plant and animal kingdoms (15, 19). Nearly every possible method for encoding information in nucleic acid is exploited by viruses, from single-stranded DNA to double-stranded RNA. Each of the 83 virus families has a different replication strategy which calls for unique proteins and unique enzymes (19). For example the replication cycles of Human herpesvirus 1 (HHV-1) and Ebolavirus (EBOV) have nothing in common (Figure 1). The dsDNA HHV-1 genome encodes 73 proteins and replicates in the host nucleus where new viral genomes are encapsidated before budding through the endoplasmic reticulum and then into vesicles that will release the virion out of the cell (6,26). The EBOV ssRNA genome encodes eight proteins, replicates in the host cell cytoplasm using its own RNA-dependent RNA polymerase complex and buds directly at the plasma membrane (2). These two disparate replication cycles only illustrate the tremendous variety of viral molecular biology. As a result, it is crucial to have a clear vision of a specific virus’ biology in order to understand its genome and protein functions. Yet this information is hardly available outside academic books.
Figure 1.
Diversity of viral replication: example of Ebolavirus versus Herpesvirus. (A) Ebolavirus is a negative single stranded RNA virus which replicates in the cytoplasm. It enters target cell by endocytosis, then penetrates into the cytoplasm by low pH fusion in the endosome. The viral RNA-dependent-RNA polymerase transcribes and replicates the viral genome in cell cytoplasm. Assembly and budding occur at the plasma membrane. (B) Herpes Simplex virus is a double stranded DNA virus which enters target cell by fusion at the plasma membrane, releasing the viral capsid in the cytoplasm. The capsid is transported to the nucleus where it injects the genomic DNA into the cell's nucleus. The viral genome circularises and conducts transcription and replication. New viral capsids are assembled in the nucleus, and bud into the endogenous reticulum (ER). These new virions fuse with ER membranes to release capsids into the cell cytoplasm. A second budding occurs at cell vesicle, and new virions are eventually released by exocytosis.
To help solve this problem, The Swiss-Prot virus annotation team has developed a website dedicated to viruses: ViralZone (www.expasy.org/viralzone). The concept of this website is to link specific knowledge for each virus family with viral protein and genomic sequences. All the available information is presented in a concise and accessible virus fact sheet. The fact sheets contain condensed information about genome, replication cycle, taxonomy and epidemiology as well as graphics describing virion organization, genome transcription and translation strategies. The whole site comprises 426 fact sheets covering the whole known virosphere: 83 families, 334 genera and nine additional pages dedicated to important species like Influenza H1N1 or HIV-1.
VIRUS TAXONOMY
Unlike Luca for cellular organism (14), there is no presumed common ancestor for viruses (12). Therefore current virus classification comprises seven independent classes, according to the Baltimore system (4). This classification is based on the nature of the nucleic acids in the virion particle: dsDNA, ssDNA, dsRNA, ss(+)RNA, ss(−)RNA, ssRNA(RT) or ssDNA(RT).
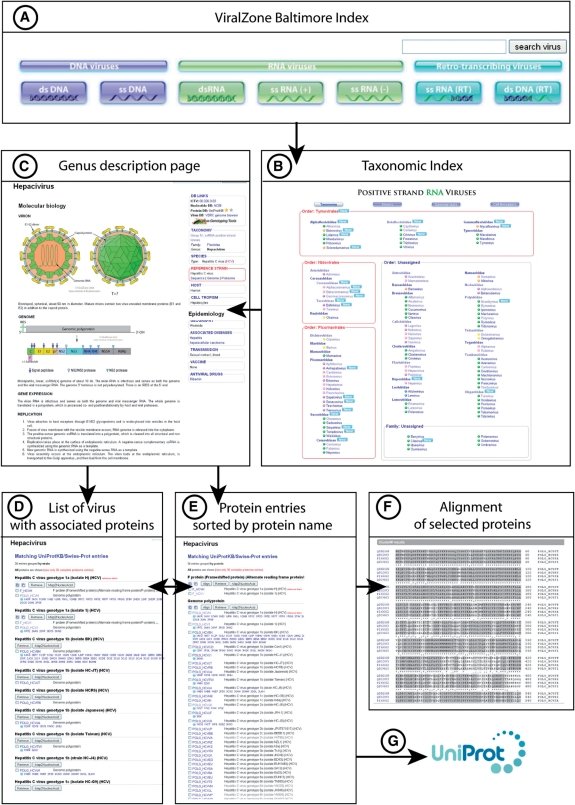
Virus abundance on earth is higher than initially expected and recent studies have unveiled millions of viruses per millilitre of seawater and billions per cubic centimeter in nearshore surface sediments (24,16); most of them are unidentified. As virus discovery accelerates, virus taxonomy has to be modified and completed each year (Table 1). In ViralZone, the starting point to access virus fact sheets are the seven Baltimore taxonomic pages (4) containing the whole list of known virus families and genera (Figure 2A). This list is reviewed each year as new viruses are constantly being described (8). The advantage of a website is that it can be incrementally updated while it can take years to publish new reference books. For example, the International Committee on Taxonomy of Viruses (ICTV) published important taxonomic changes on August 2009 on its website and the ViralZone taxonomy was updated accordingly only one month later.
Table 1.
The growing number of virus taxons
| Year | Orders | Families | Genera | Reference |
|---|---|---|---|---|
| 1995 | 2 | 54 | 184 | ICTV 6th report (20) |
| 2000 | 3 | 63 | 240 | ICTV 7th report (9) |
| 2005 | 3 | 71 | 282 | ICTV 8th report (10) |
| 2009 | 6 | 84 | 333 | ICTV online, www.ictvonline.org |
Figure 2.
(A) ViralZone Baltimore Index. (B) Taxonomic index for ssRNA(+) viruses, classified by order, family then genus. Colour spots indicate the host infected by each virus genera: pink for human and other vertebrates, purple for non-human vertebrates, green for plants, yellow for invertebrates, orange for eukaryotic microorganism, and blue for prokaryotes; (C) Genus fact sheet. (D) List of viruses referenced in UniProtKB/Swiss-Prot along with correspondent protein entries displayed by default under the fact sheet. (E) The list of entries sorted by protein names. (F) Alignment obtained after selection of protein entries in (E). (G) Each Swiss-Prot protein entry gives a direct link to UniProt web site to access to the full details of protein annotation.
From a public health point of view, providing comprehensive knowledge for all known virus genera turns out to be extremely useful when a new pathogen emerges out of a neglected virus family. A recent example is provided by the Xenotropic Moloney murine leukaemia virus-Related Virus (XMRV), which has recently raised the interest of the scientific community for its potential involvement in prostate cancer (22) and/or chronic fatigue syndrome (18). Since specific gammaretrovirus resources on the web were scarce, a direct consequence has been a dramatic increase in the number of hits to the corresponding ViralZone page (www.expasy.org/viralzone/all_by_species/67.html) that reached close to 2000 visitors in November–December 2009 (source: Google Analytics).
HOSTS
Virus host ranges can be quite narrow, e.g. the human hepatitis B virus which is strictly restricted to Human, or very large e.g. the rabies virus which seems to be able to successfully infect any mammal. Knowing the host tropism is essential to understanding the viral molecular biology. For example a dsDNA viral genome are transcribed differently in a bacteria or in a eukaryote. Moreover virus host range has a dramatic importance for public health, as illustrated by zoonosis like SARS, Ebola or Influenza that are caused by viruses able to mutate and cross hosts barriers, thus threatening the human population. For all these reasons the display of virus host tropism is highlighted in ViralZone. The hosts are indicated by a colour code for each virus genera on the taxonomy pages (Figure 2B). ViralZone display of hosts is restricted to the natural reservoirs. Vectors hosts, dead-end or laboratory hosts are not described here except if a human host/cell-line is involved.
Virus families can be browsed ‘by host’, allowing users to easily identify which viral families infect Humans, non-human vertebrates, plants, eukaryotic microorganisms, archaea or bacteria. A complete list of all major virus species able to infect humans is accessible through the ViralZone home page (www.expasy.org/viralzone/all_by_species/678.html).
GENUS AND FAMILY VIRUS FACT SHEETS
Virus fact sheets provide concise and specific information on molecular biology, taxonomy, hosts, and epidemiological data (Figure 2C). The first tab: ‘General’ describes molecular biology, virion and genome organization, followed by a step-by-step description of the viral cell infection cycle. The database section allows easy access to NCBI nucleotide and UniProtKB protein entries, as well as to specific virology databases such as VIPR (http://www.viprbrc.org) VIPERdb (7,23), Descriptions of Plant Viruses (1) and VBRC (www.vbrc.org, virology.ca). Host and cell tropism are generally indicated, but the latter might be absent since this kind of data can be unknown, or difficult to access in the literature. Cell receptor(s) for virus entry are also listed and links are provided to relevant publication(s). Finally epidemiological data briefly describe associated diseases and virus transmission as well as vaccines (if available) or antiviral drugs effective against this virus.
Under the ‘Proteins by Strain’ tab, strains and/or isolates are listed together with the proteins they encode. This list displays all related UniProtKB/Swiss-Prot entries (Figure 2D). These are manually curated entries with data extracted from publications. All the proteins annotated for a given virus strain or isolate are accessible at once for a given virus. Alternatively, the ‘Proteins by Name’ tab displays clusters of proteins having the same name and function (Figure 2E). This sorting is possible because the protein entries have been manually curated and have a coherent naming system. Calling the ClustalW alignment software (Figure 2F) directly from this page allows the user to align a set of these proteins to quickly generate a general alignment of any given viral protein family. Reference strain entries are clearly indicated, giving a landmark to users looking for the optimal data for a given viral protein family.
VIRION PICTURES
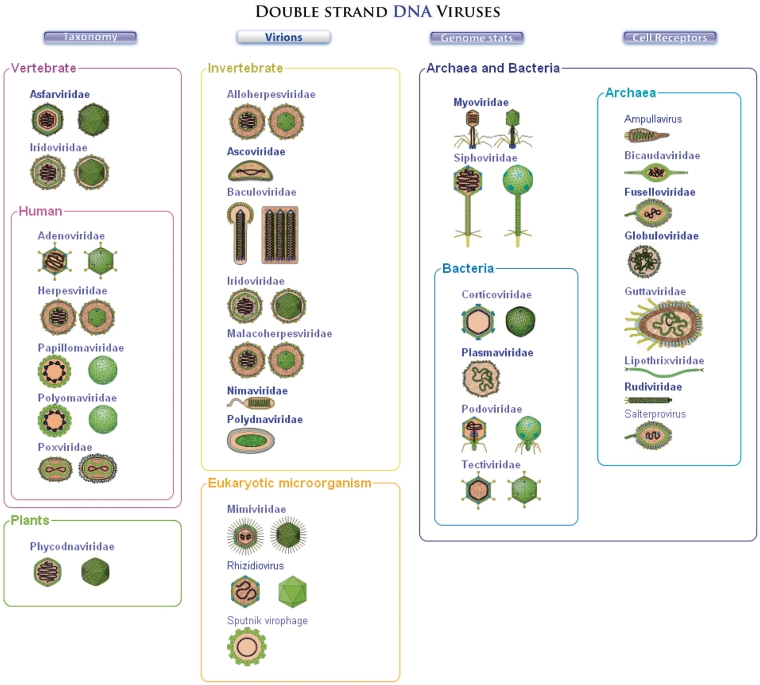
Virions are very diverse in shape and structure; they can be enveloped with one to several lipid bilayers or naked, and the genetic material can be protected by one, two or even three capsids showing helical or icosahedric symmetry and whose size is often related to genome length: from 17 nm (Porcine circovirus, 1.7 kb) to 400 nm in diameter (Mimivirus, 1200 kb). Virion pictures and diagrams can be found in Virology books or publications, but often with heterogeneous quality, colours and resolution. We created 160 original virion diagrams for ViralZone covering all known viral families and genera described to date. All the figures share the same concept and resolution, with defined colours for each part of the viral particle. Virions with icosahedric capsid symmetry are represented first as seen from a cross-section, then with 3D-like picture showing precise capsid architecture (Figure 3). Structural proteins are coloured in the same way in virion and genome pictures.
Figure 3.
This page of ViralZone displays small virion picture for all dsDNA viruses (www.expasy.org/viralzone/all_by_species/748.html). Clicking on virus family or orphan genus name gives access to the virus description page, with full size picture of virion.
All these pictures are available to the scientific community, and freely accessible on the ViralZone web site. Permission is granted to download and use them for any academic purposes: thesis, presentations or publications, provided the source is acknowledged (source: ViralZone www.expasy.ch/viralzone, Swiss Institute of Bioinformatics).
REFERENCE STRAINS: COPING WITH THE EXPONENTIAL INCREASE OF VIRUS SEQUENCES
Virus genomes are relatively small, mostly <50 kb, and are therefore easy and relatively inexpensive to sequence. This has resulted in an exponential increase in the number of new virus isolates deposited in the sequence databases. Of the 851 503 viral protein entries in UniProtKB, the species with the greatest number of open reading frames (ORF) deposited in UniProtKB is HIV-1 with 313 532 different ORFs, while the human proteome only accounts for 77 225 entries (Table 2) (UniProt release 15.12).
Table 2.
Most represented species in UniProtKB (release 15.12)
| Position | ORFs | Species | Prot/genome | Complete genome equivalents |
|---|---|---|---|---|
| 1 | 313 532 | Human immunodeficiency virus 1 | 9 | 34 329 |
| 2 | 113 396 | Influenza A virus | 11 | 10 309 |
| 3 | 95 799 | Oryza sativa subsp. japonica (Rice) | 40 577 | 2 |
| 4 | 77 225 | Homo sapiens (Human) | 20 500 | 4 |
| … | … | … | … | |
| 6 | 74 067 | Hepatitis C virus | 2 | 37 034 |
| … | … | … | … | |
| 17 | 34 367 | Hepatitis B virus (HBV) | 5 | 6873 |
As manually annotating all UniProtKB viral proteins is not achievable, we selected about one reference strain (RefStrain) per genus to be fully curated. These RefStrains have been preferentially chosen in the genus type species, and belong to the NCBI Reference Sequences database (RefSeq) in which viral genomes have been manually reviewed (5). The 355 RefStrains selected account for 12 576 proteins, which are representative of the diversity of all virus genera and can be reasonably easily maintained in an annotated and updated form to reflect ongoing research. These RefStrains are now accessible through each ViralZone fact sheet which provides links to the corresponding RefSeq genome and UniProtKB virus proteome. RefStrains allow users to know which sequences to look at in order to have the best and most up-to-date information for any given virus, and can serve as templates to correctly annotate all similar viruses, an area of high interest to the bioinformatics community.
FUTURE DIRECTIONS
ViralZone is regularly updated with new information extracted from publications and scientific meetings abstracts. Users are also actively contributing by sending feedback, minor corrections and ideas to viralzone@isb-sib.ch. Future improvements will permit the further development of the viral molecular biology section, which will in turn be linked from fact sheets. Replication cycles are for the moment described in text format for all virus fact sheets but pictures would be more suited. An example of such picture is already accessible for the Inoviridae family (www.expasy.org/viralzone/all_by_species/675.html). The design of a virus specific controlled Gene Ontology (GO) that will both facilitate gene analysis and enhance data exchange between viral sequence databases is under way in collaboration with the GO consortium (13,27).
CONCLUSION
ViralZone is a freely accessible web resource that offers accurate and concise virus information for all known viruses. It displays high quality virion pictures available to all the scientific community. The site also functions as a hub for all scientists interested in virus knowledge, by bringing together virus metadata with genomic and protein sequence databases. Indeed the ViralZone web resource has already been cited as a source of data in several publications (3,17,21,28), dozens of thesis, many scientific web sites, and the virion figures are already widely used to support communication and teaching in Virology.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Swiss Federal Government through the Federal Office of Education; Science grant Swiss Institute of Bioinformatics (SIB) (www.isb-sib.ch). Funding for open access charge: Swiss Institute of Bioinformatics.
Conflict of interest statement. None declared.
REFERENCES
- 1.Adams MJ, Antoniw JF. DPVweb: a comprehensive database of plant and fungal virus genes and genomes. Nucleic Acids Res. 2006;34(Database issue):D382–D385. doi: 10.1093/nar/gkj023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ascenzi P, Bocedi A, Heptonstall J, Capobianchi MR, Di Caro A, Mastrangelo E, Bolognesi M, Ippolito G. Ebolavirus and Marburgvirus: insight the Filoviridae family. Mol. Aspects Med. 2008;29:151–185. doi: 10.1016/j.mam.2007.09.005. [DOI] [PubMed] [Google Scholar]
- 3.Bahir I, Fromer M, Prat Y, Linial M. Viral adaptation to host: a proteome-based analysis of codon usage and amino acid preferences. Mol. Syst. Biol. 2009;5:311. doi: 10.1038/msb.2009.71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Baltimore D. Expression of animal virus genomes. Bacteriol. Rev. 1971;35:235–241. doi: 10.1128/br.35.3.235-241.1971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bao Y, Federhen S, Leipe D, Pham V, Resenchuk S, Rozanov M, Tatusov R, Tatusova T. National center for biotechnology information viral genomes project. J. Virol. 2004;78:7291–7298. doi: 10.1128/JVI.78.14.7291-7298.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Boehmer PE, Nimonkar AV. Herpes virus replication. IUBMB Life. 2003;55:13–22. doi: 10.1080/1521654031000070645. [DOI] [PubMed] [Google Scholar]
- 7.Carrillo-Tripp M, Shepherd CM, Borelli IA, Venkataraman S, Lander G, Natarajan P, Johnson JE, Brooks CL, 3rd, Reddy VS. VIPERdb2: an enhanced and web API enabled relational database for structural virology. Nucleic Acids Res. 2009;37(Database issue):D436–D442. doi: 10.1093/nar/gkn840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Carstens EB, Ball LA. Ratification vote on taxonomic proposals to the International Committee on Taxonomy of Viruses (2008) Arch. Virol. 2009;154:1181–1188. doi: 10.1007/s00705-009-0400-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fauquet CM, Mayo MA. The 7th ICTV report. Arch. Virol. 2001;146:189–194. doi: 10.1007/s007050170203. [DOI] [PubMed] [Google Scholar]
- 10.Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA. Virus Taxonomy. Classification and Nomenclature of Viruses. 2005. 8th ICTV Report, Academic Press, Elsevier. [Google Scholar]
- 11.Finsterbusch T, Mankertz A. Porcine circoviruses–small but powerful. Virus Res. 2009;143:177–183. doi: 10.1016/j.virusres.2009.02.009. [DOI] [PubMed] [Google Scholar]
- 12.Forterre P. The origin of viruses and their possible roles in major evolutionary transitions. Virus Res. 2006;117:5–16. doi: 10.1016/j.virusres.2006.01.010. [DOI] [PubMed] [Google Scholar]
- 13.Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38(Database issue):D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glansdorff N, Xu Y, Labedan B. The last universal common ancestor: emergence, constitution and genetic legacy of an elusive forerunner. Biol Direct. 2008;3:29. doi: 10.1186/1745-6150-3-29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koonin EV. On the origin of cells and viruses: primordial virus world scenario. Ann. NY Acad. Sci. 2009;1178:47–64. doi: 10.1111/j.1749-6632.2009.04992.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kristensen DM, Mushegian AR, Dolja VV, Koonin EV. New dimensions of the virus world discovered through metagenomics. Trends Microbiol. 2009;18:11–19. doi: 10.1016/j.tim.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Liechti R, Gleizes A, Kuznetsov D, Bougueleret L, Le Mercier P, Bairoch A, Xenarios I. OpenFluDB, a database for human and animal influenza virus. Database. 2010 doi: 10.1093/database/baq004. doi: 10.1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lombardi VC, Ruscetti FW, Das Gupta J, Pfost MA, Hagen KS, Peterson DL, Ruscetti SK, Bagni RK, Petrow-Sadowski C, Gold B, et al. Detection of an infectious retrovirus, XMRV, in blood cells of patients with chronic fatigue syndrome. Science. 2009;326:585–589. doi: 10.1126/science.1179052. [DOI] [PubMed] [Google Scholar]
- 19.Macnaughton TB, Lai MM. HDV RNA replication: ancient relic or primer? Curr. Top Microbiol. Immunol. 2006;307:25–45. doi: 10.1007/3-540-29802-9_2. [DOI] [PubMed] [Google Scholar]
- 20.Murphy FA, Fauquet CM, Bishop DHL, Ghabrial SA, Jarvis AW, Martelli GP, Mayo MA, Summers MD. Virus Taxonomy. Classification and Nomenclature of Viruses. Sixth Report of the International Committee on Taxonomy of Viruses. Wien New York: Springer; 1995. [Google Scholar]
- 21.Saxena SK, Mishra N, Saxena R, Saxena S. Swine flu: influenza A/H1N1 2009: the unseen and unsaid. Future Microbiol. 2009;4:945–947. doi: 10.2217/fmb.09.71. [DOI] [PubMed] [Google Scholar]
- 22.Schlaberg R, Choe DJ, Brown KR, Thaker HM, Singh IR. XMRV is present in malignant prostatic epithelium and is associated with prostate cancer, especially high-grade tumors. Proc Natl Acad Sci USA. 2009;106:16351–16356. doi: 10.1073/pnas.0906922106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 23.Shepherd CM, Borelli IA, Lander G, Natarajan P, Siddavanahalli V, Bajaj C, Johnson JE, Brooks CL, 3rd, Reddy VS. VIPERdb: a relational database for structural virology. Nucleic Acids Res. 2006;34(Database issue):D386–D389. doi: 10.1093/nar/gkj032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Suttle CA. Viruses in the sea. Nature. 2005;437:356–361. doi: 10.1038/nature04160. [DOI] [PubMed] [Google Scholar]
- 25.Suttle CA. Marine viruses–major players in the global ecosystem. Nat. Rev. Microbiol. 2007;5:801–812. doi: 10.1038/nrmicro1750. [DOI] [PubMed] [Google Scholar]
- 26.Taylor TJ, Brockman MA, McNamee EE, Knipe DM. Herpes simplex virus. Front Biosci. 2002;7:D752–D764. doi: 10.2741/taylor. [DOI] [PubMed] [Google Scholar]
- 27.The Gene Ontology Consortium. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Van Den Wollenberg DJ, Van Den Hengel SK, Dautzenberg IJ, Kranenburg O, Hoeben RC. Modification of mammalian reoviruses for use as oncolytic agents. Expert Opin. Biol. Ther. 2009;9:1509–1520. doi: 10.1517/14712590903307370. [DOI] [PubMed] [Google Scholar]