Abstract
The Protein Ontology (PRO) provides a formal, logically-based classification of specific protein classes including structured representations of protein isoforms, variants and modified forms. Initially focused on proteins found in human, mouse and Escherichia coli, PRO now includes representations of protein complexes. The PRO Consortium works in concert with the developers of other biomedical ontologies and protein knowledge bases to provide the ability to formally organize and integrate representations of precise protein forms so as to enhance accessibility to results of protein research. PRO (http://pir.georgetown.edu/pro) is part of the Open Biomedical Ontology Foundry.
INTRODUCTION
Biomedical ontologies have emerged as important tools for aggregating and integrating information to facilitate knowledge discovery in heterogeneous, genome-scale data sets (1). Ontologies are designed to make logically coherent connections in ways that computers can exploit for multiple different purposes, including data retrieval, consistency checking, integration and reasoning. In this sense they are resources that add to the power of traditional databases. The Gene Ontology (GO) (2), in particular, is a widely-used standard for gene product annotation that is now used as the basis for many different sorts of statistical analyses of biological and clinical data. Numerous ontologies covering a variety of biomedical domains are currently available, and a subset of ontology developers have committed to a collaborative effort called the Open Biomedical Ontologies (OBO) Foundry (3) to further these efforts in a systematic way.
We describe here the Protein Ontology (PRO). The PRO resource leverages and adds value to existing protein sequence resources such as UniProtKB (4) by providing an ontological representation of proteins and protein complexes. It presents a way to refer to these entities, provides rigorous definitions of these terms, and gives information concerning the relations between them—for example subtype and derivation—along the lines described in (5). Notably, PRO provides the indicated information especially with respect to the states that proteins and protein complexes can exhibit as a result of various biological processes.
THE PRO
PRO within the OBO foundry
OBO Foundry ontologies are organized along two dimensions: (i) granularity (from molecules to populations); and (ii) relation to time (objects, qualities, processes). As an OBO Foundry member, PRO is charged with covering the domain of protein-related objects at the molecular level of granularity. Specifically, PRO represents those naturally-occurring molecules that contain amino acid chains, which is to say: proteins, peptides produced by cleavage of longer amino acid chains, and protein-containing complexes from specific taxa. When appropriate, the terms in PRO are defined via cross-products using other ontology or database resources. For example, we define mature forms of leucoagglutinating phytohemagglutinin using the Sequence Ontology (SO) (6) and the Protein Modification Ontology (PSI-MOD) (7) to provide terms for parts of proteins removed during maturation and modified amino acid residues, respectively. The Chemical Entities of Biological Interest Ontology (ChEBI) (8) likewise provides terms that are used to define those PRO objects that contain non-protein attachments or components. Finally, Pfam (9) provides terms used to describe protein regions. Similarly, PRO uses terms from GO when providing relation information, for example, to the effect that a given protein or complex has a disposition toward performing some function during some process within some cellular location.
PRO meta-structure: sub-ontologies, relations and categories
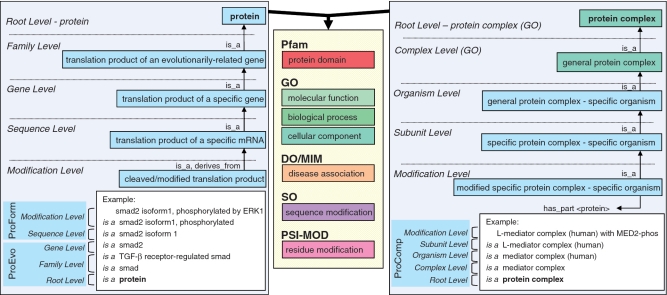
PRO encompasses three sub-ontologies: proteins based on evolutionary relatedness (ProEvo); protein forms produced from a given gene locus (ProForm); and protein-containing complexes (ProComp) (Figure 1).
Figure 1.
Overview of PRO. The left panel shows the ProEvo and ProForm subontologies, the right panel shows the ProComp subontology and the central panel shows the typical resources used to define or annotate PRO terms.
The ProEvo sub-ontology represents proteins translated from different but related (ortholog or paralog) genes. The relation used in the ProEvo hierarchy is exclusively is_a, as in ‘nodal protein is_a TGF-β-like cystine-knot cytokine’; which is to say that nodal protein is a subtype of TGF-β-like cystine-knot cytokine.
The ProForm sub-ontology represents all forms of proteins encoded by a single gene, where ‘all forms’ here refers to every splice isoform, mutation variant and co- or post-translationally modified form (and combinations thereof) that can arise from translation from that gene in any organism (see below description of 1:1 ortholog for what is meant by ‘that gene’). ProForm uses two relations: is_a denotes the subtype relation between child and parent term, and derives_from denotes the relation between a proteolytic cleavage product and its fully-formed precursor.
The ProComp sub-ontology represents specific amino acid chain-containing complexes. The definition of such entities is composed by referencing specific single chain PRO entries within the ontology (‘complex has_part subunit’). As for ProForm, there are separate terms for different versions of a complex, be that difference due to some change in modification status of one or more components or a change in the identity or stoichiometry of the components themselves. Thus, a three subunit complex where one subunit is represented by two modified forms will have two separate ProComp terms that differ only in the form for that one subunit. Like ProEvo, ProComp uses only the is_a relation in the hierarchy.
PRO terms are labeled with categories that reflect the overall organization of the ontology. Terms can be labeled as complexes and as organism-specific, as appropriate, and there are several categories used to reflect the is_a hierarchy from ProEvo to ProForm:
Family-level
PRO terms in this category refer to protein products of a distinct gene family arising from a common ancestor. The leaf-most nodes at this level are usually families comprising paralogous sets of gene products (of one or more organisms). For example, SMAD2 and SMAD3 both encode proteins belonging to the TGF-β receptor-regulated smad family, while SMAD1, SMAD5 and SMAD9 are members of the BMP receptor-regulated smad family. Thus, ‘TGF-β receptor-regulated smad protein’ and ‘BMP receptor-regulated smad protein’ are terms denoting distinct families in PRO. The family level category refers to such groupings at any degree of sequence similarity. For example, the two families indicated above can be merged into (become subclasses of) a ‘receptor-regulated smad protein’ class and further merge (with the protein products of SMAD4, SMAD6 and SMAD7) into a ‘smad protein’ class. Merging occurs only if the entire, full-length protein is evolutionarily related. Thus, for example, the smad proteins would not be found classed with nuclear factor 1 proteins even though they share one domain (MH1; Pfam:PF03165).
Gene-level
PRO terms in this category refer to the protein products of a distinct gene. For example, ‘SMAD2’ and ‘SMAD3’ are two different genes, even though they are paralogs; therefore, they have two different PRO entries at the gene level of distinction. The protein products of all alleles of human SMAD2 and mouse SMAD2 thus fall under this single term. Gene-level distinction is the leaf-most node of the ProEvo part of PRO.
Sequence-level
PRO terms in this category refer to those protein products that have a distinct sequence upon initial translation. The sequence differences can arise from different alleles of a given gene, from splice variants of a given RNA, or from alternative initiation and ribosomal frame-shifting during translation. One can think of this as a mature mRNA-level distinction. For example, SMAD2 encodes both a long splice form and a short splice form. The protein products from each isoform are separate PRO terms. Sequence-level distinction is the first (parent-most) node of the ProForm part of PRO.
Modification-level
PRO terms in this category refer to the protein products derived from a single mRNA species that differ because of some change (or lack thereof) that occurs after the initiation of translation (co- or post-translational). This includes differences due to cleavage and chemical changes to amino acid residues. For example, the long isoform of smad2 can either be unmodified or be post-translationally modified to contain phosphorylated residues. Modification-level terms are the leaf-most nodes of the ProForm part of PRO.
1:1 orthologs, orthologous isoforms and orthologous modified forms
Each gene-level term in PRO generally includes all translation products from a given gene in any organism that contains that gene. That is, a gene-level term in PRO such as ‘amyloid beta A4 protein’ refers to that same protein in human, mouse and any other organism that contains an orthologous gene. The general definition for gene-level terms conforms to the format ‘X is a protein that is a translation product of the X gene or a 1:1 ortholog thereof.’ The definition of 1:1 ortholog comes from the Homology Ontology (HOM) (10), which states that 1:1 orthology ‘… involves two genes that did not experience any duplication after the speciation event that created them.’
Special mention must be made of the way in which specific isoforms of a given gene are treated in PRO. PRO considers a protein an isoform when its primary sequence differs from that of another protein encoded by the ‘same gene’ in the ‘same organism’ due to alternative splicing or start site selection (a protein that differs due to genetic differences is termed ‘sequence variant’). As for gene-level terms, isoform terms are generally considered taxon-independent unless otherwise indicated. How then, can a given isoform term be taxon-independent (being, as they are, defined in many cases by specific sequence)? We define such orthologous isoforms as isoforms—encoded by orthologous genes—that are believed to have arisen prior to speciation and divergence of the primary sequence. That is, the extant orthologous isoforms existed as a single isoform in a common ancestor. Thus if the human and mouse isoforms are deemed to be orthologous, a single PRO term unifying them is created, and the database entries providing the sequence for each is mapped to the single PRO term.
PRO also contains separate entries for specific modified forms of a protein that may exist in multiple organisms. For example, human c-myc protein (UniProtKB:P01106) can be phosphorylated at positions 58T, 62S and 71S (among others) (11). PRO contains explicit entries for the 62Sphos form, the 58Tphos+62Sphos form and the 62Sphos+71Sphos form. PRO would not contain a 58Tphos+71Sphos form until such is observed. The 58Tphos+62Sphos form of c-myc includes c-myc in any organism that contains phosphoresidues corresponding to those indicated. Thus, the PRO term for c-myc 58Tphos+62Sphos includes both human c-myc 58Tphos+62Sphos and cottontail rabbit c-myc (UniProtKB:Q9MZT6) 59Tphos+63Tphos, among others.
PRO DATA
Sources and content
PRO information is obtained by manual curation from the scientific literature and by large-scale processing of resources that provide curated protein and pathway information. Predominant among these, especially for gene-level terms in PRO, is UniProtKB. The information from this resource includes protein name and synonyms and, indirectly, the definition. Term name and definition follow the conventions used for the human protein. If no human entry exists, the name is taken from the mouse or Escherichia coli protein. Indications of initiator methionine removal or signal peptide processing (for example) also come from this resource. Terms invoking residue modifications come from manual curation, or from other sources that explicitly indicate how multiple possible modifications combine. These include Reactome (12), MouseCyc (13) and EcoCyc (14). These resources describe both precise proteins forms and precise protein complexes. Both types of entities are thus being imported into PRO on a large-scale basis with curator monitoring. The main sources of terms in the ProEvo sub-ontology are PIRSF (15) and Panther (16). PRO term definitions can be found in the OBO-format file. Information for each entry is given in a stanza that provides term ID, name, definition, synonyms, category, entry status (if obsolete) and relationship to other terms.
Annotation
Curators inspect the scientific literature to find references to protein forms and protein complexes described therein and to identify associated evidence. Specific attributes (such as protein function, localization, process and involvement in disease) are included in the PRO association file (PAF). All annotations are formulated using terms from controlled vocabularies [GO, SO, PSI-MOD, MIM (17), ChEBI, Pfam] and include the corresponding relation.
Database cross-references
To facilitate navigation from PRO to other resources that provide information about protein-related entities, we provide a downloadable tab-delimited list of cross-references (promapping.txt). The file explicitly indicates how the mapped entity relates to the PRO term, where is_a means that the mapped entity is a subtype of the PRO entity and ‘exact’ means that they denote the same entity. Currently the mapping file contains cross-references to UniProtKB, HGNC (18), MGI (19), EcoCyc and Reactome for human, mouse and E. coli proteins. In the future the mapping will contain all twelve organisms included in the GO Reference Genome Annotation project (20).
Data coverage
As stated above, PRO will eventually represent proteins from the 12 GO Reference Genomes. The number of possible protein forms within this set is as yet unknown, but we can make a good estimate of the maximum number of gene level terms using predicted orthologs from Inparanoid 7 (21). Out of the nearly 200 000 species-specific genes, we estimate a total of ∼100 000 gene-level, 1:1 ortholog-based terms. It is similarly not possible to know how many complexes will eventually be identified and represented in PRO, though for example Reactome has ∼2700 complexes. Progress toward these goals can be monitored from the statistics page (http://pir.georgetown.edu/cgi-bin/pro/sta_pro).
ISSUES CONCERNING PRO DEVELOPMENT
Speed of curating terms
Manual curation is needed to establish a high-quality core data set and to develop standards for continued growth. Not surprisingly, proper enumeration of forms, complexes and variants thereof is laborious, so getting good coverage and good quality is a challenge. Collaborating with other resources to capture large blocks of data is one approach to providing content, and furthermore is a useful framework for developing tools and standards for data importation. We are piloting the use of BioPAX-formatted pathway information to extract terms for complexes and protein forms from Reactome, a process which will then be extensible to other BioPAX-formatted resources.
Data representation
As illustrated by the c-myc example above, orthologous modified forms present a challenge for formal representation because the taxon-specific children of such terms do not necessarily share the same sequence or start position. Such terms therefore cannot be defined by referring to specific positions. We currently define them using free text and either an example sequence or a motif, but are exploring ways to formalize these.
COMMUNITY ACCESS
Web access and browsing
The PRO homepage (http://pir.georgetown.edu/pro) is the starting point for navigation through the PRO resources, providing both quick and advanced term and annotation searches, links to associated documents and background information pages, an ftp download page, term request and submission pages, and an ontology browser. Advanced search, quick browse and result pages offer the ability to refine the output based on predefined topics that are of special interest to biologists, including (i) by modified protein form (any modification, or specific types, such as phosphorylation or acetylation); (ii) by complex; (iii) clinical-related terms such as saliva biomarkers; or (iv) those with link to an external database. In addition, Boolean (AND, OR, NOT) searches are supported, as are ‘null’ (not present) and ‘not null’ (present). Results are shown in a table format and the columns displayed can be customized. The table can be saved as a tab-delimited file. In addition, PRO terms can be selected to view the hierarchy in the browser.
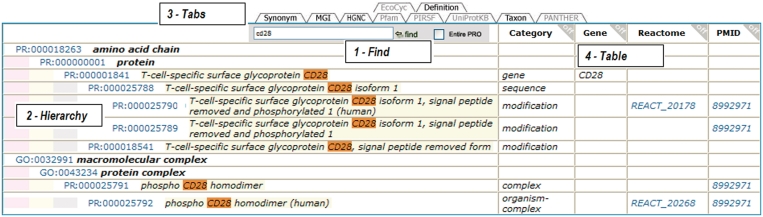
The browser displays the structure of the ontology and overlays information available for displayed terms. It has the capability to show the complete ontology by clicking ‘Browser’ in the PRO homepage, or to display the hierarchy for selected terms by selecting PRO terms in the result table or by selecting the icon next to the PRO term. Within the Browser page, the ‘find’ functionality displays terms matching a given word or phrase. In addition, customized data can be displayed by selecting the specific tabs (Figure 2).
Figure 2.
PRO browser displaying the hierarchy for CD28-containing terms (partial view). (1) The Find box provides a means to display or highlight terms that match a specific word or phrase. (2) The hierarchy is displayed with term highlighted. (3) Information tabs: click on a black tab to display the information in the table (grayed-out tabs either do not apply or have already been selected for display). (4) Customizable table of selected information. In the example shown Gene, Reactome cross-reference and PMID were selected. Category is shown by default.
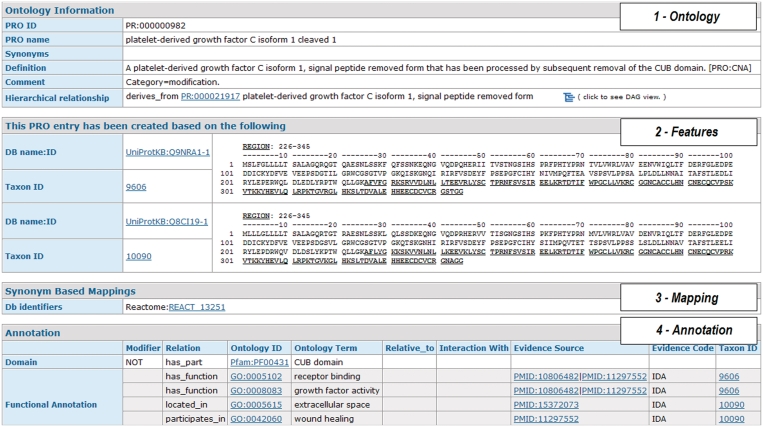
Clicking on a PRO ID from any page brings up the corresponding entry report. The report combines the information from the ontology file, the annotation file and the mapping file. It provides a graphical display of the sequences including modifications plus links to external resources (Figure 3). The entry report is divided into four parts: (i) Ontology information, showing the definition of the protein or complex object, and linking to the PRO hierarchy for this term. (ii) The representative proteins for a given protein form with experimental evidence. Figure 3 shows one example, a proteolytic cleavage product of isoform 1 of platelet derived growth factor C. This form is conserved in human and mouse, and therefore two sequences are displayed along with the accompanying features; in this case the proteolytic cleavage product (underlined; region 226–345 in both organisms). (iii) Mapping to identical objects in an external database. In this example the link is to Reactome, which has this protein entity because it is involved in a biological pathway. (iv) The annotation for the particular protein form (or complex) described in the entry expressed via relationships to other ontologies and/or databases.
Figure 3.
Sample PRO entry report for a cleaved form of platelet-derived growth factor C (partial view). (1) Ontology section, with ID, name and synonyms, definition, comments and hierarchy links. (2) Sequence and features for representative entities. For cleaved forms, the underlining indicates the entity of interest while red letters (not shown in this example) indicate modified residues. (3) Mapped cross-references to the identical entity in an external resource. (4) The annotation section.
Documentation and data distribution
All documents and data are available from the PRO website. Documents include information about PRO annotation guidelines and standards, the description of the ontological framework, a website tutorial to help navigate the website and Q&A to address frequent questions. PRO distribution files include the ontology in OBO format (pro.obo), the accompanying annotation file (PAF.txt) in a tab delimited format, and mappings to external databases, also tab delimited.
Integration of PRO within other community database resources
Links to PRO are available from gene, protein and pathway databases such as Mouse Genome Informatics (MGI; http://www.informatics.jax.org), MouseCyc (http://mousecyc.jax.org), Reactome (http://www.reactome.org/) and EcoCyc (http://ecocyc.org), and via LinkOut from the National Center for Biotechnology Information (http://www.ncbi.nlm.nih.gov/). PRO serves as a reference ontology, for example, for the Dendritic Cell Ontology (22), The Saliva Ontology (23) and the GO (24).
Submitting and requesting PRO terms
New terms can be requested via the PRO tracker at http://sourceforge.net/tracker/?func=add&group_id=266825&atid=1135711. Requests are best submitted by referring to a UniProtKB accession and providing as much information as possible, including any relevant literature citations. Terms can also be submitted via RACE-PRO (http://pir.georgetown.edu/cgi-bin/pro/race_pro), a web-based interface that facilitates defining and annotating protein objects without requiring knowledge of ontologies or formatting restrictions. The protein object is defined based on experimental evidence from a paper describing, for example, some modification that applies to a proteolytic cleavage product. It is then possible to add annotation using a GO, Pfam, SO or MIM term. The submitted information is checked and approved by a PRO editor and converted into the appropriate format. The final product is sent back to the submitter for approval.
Linking to PRO
Terms in the PRO and associated information can be found in all cases using persistent URLs of the form http://purl.obolibrary.org/obo/PR_xxxxxxxxx where the x’s represent the numeric portion of the PRO ID. Interested parties are invited to follow PRO-related discussions and announcements by joining the PRO mailing list pro-obo-discuss@lists.sourceforge.net. There is also a wiki page that presents summary materials and information about meetings. All pages are accessible from the PRO home page.
FUNDING
National Institutes of Health (R01GM080646); the supplement grants (R01GM080646-03S1 to J.N.); (R01GM080646-04S2). Funding for open access charge: National Institutes of Health (R01GM080646).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank PRO users, workshop attendees and other interested parties who have contributed terms, advice and stimulating discussions. The authors are especially indebted to Alan Ruttenberg and Chris Mungall for many advice sessions and to Peter Karp for facilitating import of EcoCyc data.
REFERENCES
- 1.Blake JA, Bult CJ. Beyond the data deluge. Biomedical Inform. 2006;39:314–320. doi: 10.1016/j.jbi.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 2.The Gene Ontology Consortium. The Gene Ontology: extensions and refinements. Nucleic Acids Res. 2010;38(Database issue):D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith B, Ashburner M, Rosse C, Bard J, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.UniProt Consortium. The universal protein resource (UniProt) in 2010. Nucleic Acids Res. 2010;38(Database issue):D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith B, Ceusters W, Klagger B, Kohler J, Kumar A, Lomax J, Mungall C, Neuhaus F, Rector AL, Rosse C. Relations in Biomedical Ontologies. Genome Biol. 2005;6:R46. doi: 10.1186/gb-2005-6-5-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Mungall CJ, Batchelor C, Eilbeck K. Evolution of the sequence ontology terms and relationships. J. Biomed. Inform. 2010 doi: 10.1016/j.jbi.2010.03.002. doi:10.1016/j.jbi.2010.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Montecchi-Palazzi L, Beavis R, Binz PA, Chalkley RJ, Cottrell J, Creasy D, Shofstahl J, Seymour SL, Garavelli JS. The PSI-MOD community standard for representation of protein modification data. Nat. Biotechnol. 2008;26:864–866. doi: 10.1038/nbt0808-864. [DOI] [PubMed] [Google Scholar]
- 8.de Matos P, Alcántara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical entities of biological interest: an update. Nucleic Acids Res. 2010;38(Database issue):D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38(Database issue):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Roux J, Robinson-Rechavi M. An ontology to clarify homology-related concepts. Trends Genet. 2010;26:99–102. doi: 10.1016/j.tig.2009.12.012. [DOI] [PubMed] [Google Scholar]
- 11.Dephoure N, Zhou C, Villén J, Beausoleil SA, Bakalarski CE, Elledge SJ, Gygi SP. A quantitative atlas of mitotic phosphorylation. Proc. Natl Acad. Sci. USA. 2008;105:10762–10767. doi: 10.1073/pnas.0805139105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37(Database issue):D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Evsikov AV, Dolan ME, Genrich MP, Patek E, Bult CJ. MouseCyc: a curated biochemical pathways database for the laboratory mouse. Genome Biol. 2009;10:R84. doi: 10.1186/gb-2009-10-8-r84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Keseler IM, Bonavides-Martínez C, Collado-Vides J, Gama-Castro S, Gunsalus RP, Johnson DA, Krummenacker M, Nolan LM, Paley S, Paulsen IT, et al. EcoCyc: a comprehensive view of Escherichia coli biology. Nucleic Acids Res. 2009;37(Database issue):D464–D470. doi: 10.1093/nar/gkn751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nikolskaya AN, Arighi CN, Huang H, Barker WC, Wu CH. PIRSF family classification system for protein functional and evolutionary analysis. Evol. Bioinform. Online. 2007;2:197–209. [PMC free article] [PubMed] [Google Scholar]
- 16.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38(Database issue):D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Amberger J, Bocchini CA, Scott AF, Hamosh A. McKusick's Online Mendelian Inheritance in Man (OMIM) Nucleic Acids Res. 2009;37(Database issue):D793–D796. doi: 10.1093/nar/gkn665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bruford EA, Lush MJ, Wright MW, Sneddon TP, Povey S, Birney E. The HGNC Database in 2008: a resource for the human genome. Nucleic Acids Res. 2008;36(Database issue):D445–D448. doi: 10.1093/nar/gkm881. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bult CJ, Kadin JA, Richardson JE, Blake JA, Eppig JT Mouse Genome Database Group. The mouse genome database: enhancements and updates. Nucleic Acids Res. 2010;38(Database issue):D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Reference Genome Group of the Gene Ontology Consortium. The gene ontology's reference genome project: a unified framework for functional annotation across species. PLoS Comput. Biol. 2009;5:e1000431. doi: 10.1371/journal.pcbi.1000431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ostlund G, Schmitt T, Forslund K, Köstler T, Messina DN, Roopra S, Frings O, Sonnhammer EL. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38(Database issue):D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Masci AM, Arighi CN, Diehl AD, Lieberman AE, Mungall C, Scheuermann RH, Smith B, Cowell LG. An improved ontological representation of dendritic cells as a paradigm for all cell types. BMC Bioinformatics. 2009;10:70. doi: 10.1186/1471-2105-10-70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ai J, Smith B, David WT. Saliva ontology: an ontology-based framework for a Salivaomics Knowledge Base. BMC Bioinformatics. 2010;11:302. doi: 10.1186/1471-2105-11-302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mungall CJ, Bada M, Berardini TZ, Deegan J, Ireland A, Harris MA, Hill DP, Lomax J. Cross-product extensions of the Gene Ontology. J. Biomed. Inform. 2010 doi: 10.1016/j.jbi.2010.02.002. doi:10.1016/j.jbi.2010.02.002. [DOI] [PMC free article] [PubMed] [Google Scholar]