Abstract
T1DBase (http://www.t1dbase.org) is web platform, which supports the type 1 diabetes (T1D) community. It integrates genetic, genomic and expression data relevant to T1D research across mouse, rat and human and presents this to the user as a set of web pages and tools. This update describes the incorporation of new data sets, tools and curation efforts as well as a new website design to simplify site use. New data sets include curated summary data from four genome-wide association studies relevant to T1D, HaemAtlas—a data set and tool to query gene expression levels in haematopoietic cells and a manually curated table of human T1D susceptibility loci, incorporating genetic overlap with other related diseases. These developments will continue to support T1D research and allow easy access to large and complex T1D relevant data sets.
INTRODUCTION
T1DBase is a public resource that integrates public and private data from genomic and genetic resources relevant to the study of susceptibility of Type 1 Diabetes (TID). Data are incorporated from a variety of public data sets into a disease agnostic core database. This core database acts as a scaffold into which T1D specific data sets are incorporated. Currently T1DBase focuses on two main areas of research incorporating expression and genetic data sets. In terms of genetics, over the past decade the number of convincing human T1D susceptibility loci that have been discovered has accelerated from a handful in 2007 to a present count of more than 50 separate convincing loci (1). This has been mainly driven by genome-wide association studies that have been carried out in multiple diseases (2–5). The National Human Genome Research Institute Genome Wide Association Study (NHGRI GWAS) (6) catalogue contains most of the headline associations but it is also paramount to constantly update and curate these data in the light of new genetic and functional data. Indeed, there is significant overlap between T1D and other diseases (7) and teasing out these relationships requires a non-trivial investment in literature curation. To maximize utility, these efforts must be combined in the context of genomic and functional data. At present T1DBase integrates data from four genome-wide association studies (2–4), curated regions of T1D susceptibility in Human, Mouse (8) and Rat (9), and T1D relevant expression data sets (10,11).
T1DBase provides a platform that allows the integration of these complex and disparate data sets thereby linking large scale expression, genetics and genomics resources with support for integrated querying and visualization to enhance T1D research. Due to the generic framework (GDxBase) underlying T1DBase, this approach can be tailored for other diseases and is currently employed by Prion Disease Database (PDDB) (prion disease) and Glioblastoma Database (GBMBase) (glioblastoma). All software is available through a SourceForge repository under the GNU's Not Unix (GNU) General Public License or the Perl Artistic License. Most data sets are available for download from within the site.
SITE REDESIGN
The overall layout of T1DBase has been completely overhauled. The main reasons for this were 3-fold. First, from user feedback it became clear that although resources were available on the site they were often not intuitively available or required the user to follow a number of links to access. Second, to accommodate specific tasks it made sense to collect certain tools and resources together in a context specific manner into what are termed ‘portals’. Finally, by rationalizing site content substantial enhancements to the maintainability, reliability and performance of the platform were gained.
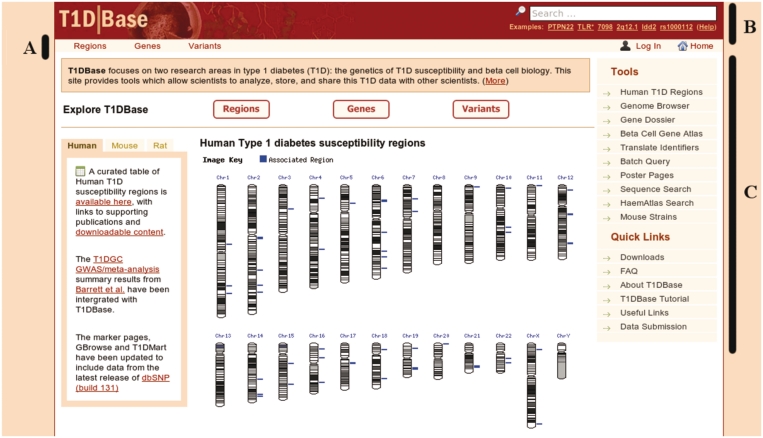
A screenshot of an example page is shown in Figure 1 and the main elements of the search bar, title bar and navigation bar are described below.
Figure 1.
Screenshot of an example T1DBase page showing the overall layout of the page. The various components are described in more detail in the text.
Title bar (A)
The title bar below the main header contains quick links to all three major ‘portals’. A portal is defined as a page that functions as a point of access to all resources based on a biological concept or idea. Each portal page is characterized by three main sections. The top ‘boilerplate’ explains context as well as drawing attention to specific source dependencies. Each portal includes a set of advanced searches that augments the top search bar and provides a single click solution to commonly asked questions. By way of example: under the region portal users can find out if a target gene of interest is located within a T1D susceptibility region previously implicated in Human, Mouse or Rat. The returned table contains links to other areas of the site that pertain to any regions retrieved. Currently three such portals are implemented on the site: Region, Gene and Variant.
Search bar (B)
The search bar is present on each page and allows a user to search underlying databases in T1DBase. Current identifiers supported include, entrez, refseq, dbSNP (rs, ss and synonyms), Online Mendelian Inheritance In Man (OMIM) and gene symbols [HGNC, mouse genome informatics (MGI) and rat genome database (RGD)]. Search has been updated to allow searching for T1D specific variant aliases (e.g. CT60) and loci (e.g. Idd5.3 and 12q13.3).
The region portal is focused on genomic regions of interest in T1D that are curated from the literature in all organisms. These include three main subtypes: associated regions—identified from genome-wide association studies, the boundary of these regions is set by utilizing method employed by Barrett et al. (2); linkage regions—mainly identified from model organism congenic mapping and orthologous regions—which are composed of orthologous regions between Human associated regions and Mouse and Rat genomes using the Compara component of Ensembl (12).
The gene portal takes a species agnostic view of genes and attempts to link human, mouse and rat genes into orthologous units using underlying data sources (12–16). For each species, we then define a span which is the maximal span that includes all transcript annotations from all gene annotation resources (12,17,18). Currently the portal integrates this information with region and variant-based information.
The variant portal integrates public human SNP variants from Ensembl, 1000 Genomes Project and dbSNP (19) (versions 130 and 131) with region- and gene-based information.
For a particular instance of the three main biological concepts covered: Genes, Variants and Regions, an ‘overview’ page that summarizes the data for each has been developed. These contain a slice through all the resources that are relevant to a concept. Within an overview page each relevant resource is represented by a modular section that gives a simplified result for an instance. For example, a region overview page will contain sections that are relevant to a genomic region, such as lists of overlapping genes, variants and supporting publications. Where possible these are linked so that a user can access more detailed information pertaining to a particular biological concept or research question.
Navigation bar (C)
The navigation bar is context specific and is subdivided into ‘Tools’ and ‘Quick Link’ sections. The latter contains links to static resources that are relevant to the current page often taking the form of ‘Poster’ pages that support a particular publication. For overview pages this is modified to a list of page sections and provides a method to quickly link to sections of interest.
NEW DATA SETS AND TOOLS
Where possible T1DBase uses tools that are already available from other open source projects: For example, GBrowse (20) is used for genome browsing and the batch query interface to underlying data sets is based on the BioMart project (21). A similar policy is adopted for most data resources. These are only integrated if they act as a scaffold to organize T1DBase specific resources or due to performance reasons where their omission would significantly impede site operation. Recent data sets incorporated include the large scale expression and genetic data sets outlined below.
Genetic
Three genome-wide association studies directly pertaining to T1D research have been integrated (2–4). T1DBase also supports one imputation-based analysis genome-wide association meta-analysis (22) and in the near future will display the imputation based on information from the 1000 Genomes Project. Summary data for all of these studies is available for each variant included through the relevant Variant overview page. For non-authorized individuals some data is hidden to prevent personally identifiable information from being released (23). Summary data displayed includes tables of association statistics, exclusions and genotype calling ‘cluster’ plots, which allow users to asses the likelihood of genotyping error as the source of association. Tools have been incorporated to allow users to locate variants and genomic regions that are in high linkage disequilibrium with currently selected variant using HapMap (24) and 1000 Genomes Project data. The underlying tool uses R (25) and specifically the snpMatrix (26) package to calculate LD statistics in real time which are then presented to the user. To augment this variant specific approach, selected data is available in batch form within the batch query tool, allowing users to download results based on coordinate information. Summary association data can be visualized using the genome browser tool where it appears as a scatter plot. Each association point links back to a variant overview page, allowing users to access detailed information on a particular association.
HaemAtlas
To complement the Beta Cell Gene Atlas tool, another transcriptomics resource examining gene expression in haematopoietic cells (11) was reanalysed and incorporated into the site. This tool allows a user to query the HaemAtlas data set and retrieve the expression profile of a gene of interest; additionally it assesses whether this gene is differentially expressed across lymphoid, myeloid and haematopoietic precursor cells. A simple graphical format shows the expression pattern (obtained from probes mapping uniquely to the gene) as a bar plot. Beneath the profile, the genomic context of mapped probes, i.e. the transcripts these probes hybridize with, is displayed utilizing a snapshot from the genome browsing tool. Analysis was performed using R, Bioconductor (27) and the LIMMA package (28), with the probe-transcript matches obtained from Ensembl.
Curation
As previously stated: with the advent of recent advances in high-throughput technologies (GWAS and resequencing), there have been an explosion in the number of genetic results relevant to human T1D and complex disease. T1DBase maintains an up-to-date table of human T1D susceptibility regions. From the literature, genetic studies are hand curated and appraised for relevance within the context of T1D using the method defined in Smyth et al. (7). If a study has identified and confirmed a novel locus or overlaps a known locus based on statistical evidence then the table is updated accordingly. Full integration of the new regions into the main T1D platform occurs at the next release. The resultant table contains a list of loci indicating cytogenetic and physical (NCBI36 and GRCh37 genome builds) location, candidate gene (if any) and index variant along with disease association statistics. The studies within a region are listed in temporal order so as to demonstrate disease providence. Where there are multiple variants within a region this indicates multiple independent effects. The tool automatically computes a list of coding and non-coding genes that overlap the defined region. To account for possible long distance functional effects, similar lists are also provided for regions proximal and distal by 0.5 MB to the defined region to account for the possibility of any local acting factors. All entries are fully referenced. The table itself is available for download as PDF or text format and a list of genes and regions is available for bulk download and use in further analysis. We include those index variants with highest disease association as a specific track within the genome browser tool.
Archive site
A straightforward archive application has been developed to overcome issues with continuity and compatibility. When there is a large change to genomic data such as a new genome build for an organism, remapping of features occurs where possible. However, coordinates in publications may well be based on previous builds and these need support. The archive site is available from archive-t1dbase.org and is integrated into the main site via links from all main pages including Variant, Gene and Region overview pages. Additionally it provides a logical location for archived data sets, software and pages that are no longer included on the site but have been referenced in publications.
Sequence-based searches
T1DBase contains a simple sequence search tool that uses the BLAT (29) software to search human and mouse T1D regions. Users submit a sequence in FASTA format and the results are then rendered as set of ideograms. Hits are represented as coloured triangles that scaled in size by the number of bases matched and by colour to represent the score (red indicating the highest scoring hit and blue a low scoring hit) Users can mouse over the hit’s to examine more closely alignment or genomic context using the genome browser tool. A tabular view of the data is also available as an alternative view of the results.
DISCUSSION AND FUTURE PLANS
T1DBase continues to be a unique resource for T1D researchers. It provides a resource for integrating genetic, genomic and functional data that is disease relevant and curated within a standard website paradigm. By way of example a user interested in a particular variant can search the site and rapidly identify whether the variant (or a variant in high linkage disequilibrium) is associated with T1D across different studies. Similarly a mouse focused researcher can interrogate the database for a gene of interest and find out from a single page if human genetic data implicates the gene in T1D, and if so what tissues the gene appears to be expressed in.
At the moment, there is a focus on convincingly supported genetic association results, although negative results are also important, and future curation efforts will address this issue, by providing information on GWAS signals that have not replicated. Current efforts in T1D genetics (e.g. ImmunoChip consortium) are to fine map human regions defined from GWA and follow up studies, and as these data become available they will be integrated into T1DBase. Currently investigations are ongoing for integration of summary data from targeted high-throughput sequencing experiments, for example, allele-specific expression in human T-cells (30).
One of the next steps in complex disease is to combine genetic and functional data in relevant tissue types to obtain new insights into disease aetiology. One example of this is association between genotype and mRNA expression levels detailed in a recent study of human monocytes (31). These data will be incorporated into an external expression Quantitative Trait Loci (eQTL) browser available via the web (32) where they will be integrated with other similar studies. T1DBase will provide integrated links to these external resources, so that researchers can easily jump to a genomic region of interest. This example demonstrates that with the sheer amount of data being generated it is important not to over extend and loose focus, and that one such way to avoid this is to provide better interoperability with external resources. Within this federated paradigm other data sets within T1DBase can be leveraged so that they are accessible to external resources and vice versa by employing standard protocols, such as Distributed Annotation System (DAS) (33), bigBed and bigWig (34).
FUNDING
The Wellcome Trust (WT); the National Institute of Diabetes and Digestive Kidney Diseases (NIDDK); the National Institute for Health Research (NIHR); Juvenile Diabetes Research Foundation (JDRF). T1DBase is hosted at the Cambridge Institute for Medical Research (CIMR). Funding for open access charge: The Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We acknowledge Nathan Goodman, Barry Healy, James Allen, Victor Cassen, Lisa Iype, Denise Maudlin, Ryan Bressler and Sutee Dee for previous contributions to the project. We also thank the members of the JDRF/WT Diabetes and Inflammation Laboratory and the Type 1 Diabetes Genetic Consortium (T1DGC) for their kind support and feedback. Systems support was provided by Vin Everett and Wojciech Giel.
REFERENCES
- 1.Todd JA. Etiology of type 1 diabetes. Immunity. 2010;32:457–467. doi: 10.1016/j.immuni.2010.04.001. [DOI] [PubMed] [Google Scholar]
- 2.Barrett JC, Clayton DG, Concannon P, Akolkar B, Cooper JD, Erlich HA, Julier C, Morahan G, Nerup J, Nierras C, et al. Genome-wide association study and meta-analysis find that over 40 loci affect risk of type 1 diabetes. Nat. Genet. 2009;41:703–707. doi: 10.1038/ng.381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cooper JD, Smyth DJ, Smiles AM, Plagnol V, Walker NM, Allen JE, Downes K, Barrett JC, Healy BC, Mychaleckyj JC, et al. Meta-analysis of genome-wide association study data identifies additional type 1 diabetes risk loci. Nat. Genet. 2008;40:1399–1401. doi: 10.1038/ng.249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Wellcome Trust Case Control Consortium. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447:661–678. doi: 10.1038/nature05911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Qu HQ, Grant SF, Bradfield JP, Kim C, Frackelton E, Hakonarson H, Polychronakos C. Association of RASGRP1 with type 1 diabetes is revealed by combined follow-up of two genome-wide studies. J. Med. Genet. 2009;46:553–554. doi: 10.1136/jmg.2009.067140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hindorff LA, Sethupathy P, Junkins HA, Ramos EM, Mehta JP, Collins FS, Manolio TA. Potential etiologic and functional implications of genome-wide association loci for human diseases and traits. Proc. Natl Acad. Sci. USA. 2009;106:9362–9367. doi: 10.1073/pnas.0903103106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Smyth DJ, Plagnol V, Walker NM, Cooper JD, Downes K, Yang JH, Howson JM, Stevens H, McManus R, Wijmenga C, et al. Shared and distinct genetic variants in type 1 diabetes and celiac disease. N. Engl. J. Med. 2008;359:2767–2777. doi: 10.1056/NEJMoa0807917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ridgway WM, Peterson LB, Todd JA, Rainbow DB, Healy B, Burren OS, Wicker LS. In: Advances in Immunology. Emil RU, Hugh OM, editors. Vol. 100. Academic Press; 2008. pp. 151–175. [DOI] [PubMed] [Google Scholar]
- 9.Wallis RH, Wang K, Marandi L, Hsieh E, Ning T, Chao GY, Sarmiento J, Paterson AD, Poussier P. Type 1 diabetes in the BB rat: a polygenic disease. Diabetes. 2009;58:1007–1017. doi: 10.2337/db08-1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kutlu B, Burdick D, Baxter D, Rasschaert J, Flamez D, Eizirik DL, Welsh N, Goodman N, Hood L. Detailed transcriptome atlas of the pancreatic beta cell. BMC Med Genomics. 2009;2:3. doi: 10.1186/1755-8794-2-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Watkins NA, Gusnanto A, de Bono B, De S, Miranda-Saavedra D, Hardie DL, Angenent WG, Attwood AP, Ellis PD, Erber W, et al. A HaemAtlas: characterizing gene expression in differentiated human blood cells. Blood. 2009;113:e1–e9. doi: 10.1182/blood-2008-06-162958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl 2009. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2007;35:D5–D12. doi: 10.1093/nar/gkl1031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Bult CJ, Eppig JT, Kadin JA, Richardson JE, Blake JA. The Mouse Genome Database (MGD): mouse biology and model systems. Nucleic Acids Res. 2008;36:D724–D728. doi: 10.1093/nar/gkm961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Twigger SN, Shimoyama M, Bromberg S, Kwitek AE, Jacob HJ. The Rat Genome Database, update 2007: easing the path from disease to data and back again. Nucleic Acids Res. 2007;35:D658–D662. doi: 10.1093/nar/gkl988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen F, Mackey AJ, Stoeckert CJ, Jr, Roos DS. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 2010;38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Pruitt KD, Harrow J, Harte RA, Wallin C, Diekhans M, Maglott DR, Searle S, Farrell CM, Loveland JE, Ruef BJ, et al. The consensus coding sequence (CCDS) project: identifying a common protein-coding gene set for the human and mouse genomes. Genome Res. 2009;19:1316–1323. doi: 10.1101/gr.080531.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Res. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Smedley D, Haider S, Ballester B, Holland R, London D, Thorisson G, Kasprzyk A. BioMart–biological queries made easy. BMC Genomics. 2009;10:22. doi: 10.1186/1471-2164-10-22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Wallace C, Smyth DJ, Maisuria-Armer M, Walker NM, Todd JA, Clayton DG. The imprinted DLK1-MEG3 gene region on chromosome 14q32.2 alters susceptibility to type 1 diabetes. Nat. Genet. 2010;42:68–71. doi: 10.1038/ng.493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Homer N, Szelinger S, Redman M, Duggan D, Tembe W, Muehling J, Pearson JV, Stephan DA, Nelson SF, Craig DW. Resolving individuals contributing trace amounts of DNA to highly complex mixtures using high-density SNP genotyping microarrays. PLoS Genet. 2008;4:e1000167. doi: 10.1371/journal.pgen.1000167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–U853. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.R Core Development Team. R: A Language and Environment for Statistical Computing. 2005. Vienna, Austria. [Google Scholar]
- 26.Clayton D, Leung HT. An R package for analysis of whole-genome association studies. Hum. Hered. 2007;64:45–51. doi: 10.1159/000101422. [DOI] [PubMed] [Google Scholar]
- 27.Gentleman RC, Carey VJ, Bates DM, Bolstad B, Dettling M, Dudoit S, Ellis B, Gautier L, Ge Y, Gentry J, et al. Bioconductor: open software development for computational biology and bioinformatics. Genome Biol. 2004;5:R80. doi: 10.1186/gb-2004-5-10-r80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Smyth GK. Limma: linear models for microarray data. In: Gentleman R, Carey V, Dudoit S, Irizarry R, Huber W, editors. Bioinformatics and Computational Biology Solutions using R and Bioconductor. New York: Springer; 2005. pp. 397–420. [Google Scholar]
- 29.Kent WJ. BLAT: the BLAST-like alignment tool. Genome Res. 2002;12:656–664. doi: 10.1101/gr.229202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Heap GA, Yang JH, Downes K, Healy BC, Hunt KA, Bockett N, Franke L, Dubois PC, Mein CA, Dobson RJ, et al. Genome-wide analysis of allelic expression imbalance in human primary cells by high-throughput transcriptome resequencing. Hum. Mol. Genet. 2010;19:122–134. doi: 10.1093/hmg/ddp473. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zeller T, Wild P, Szymczak S, Rotival M, Schillert A, Castagne R, Maouche S, Germain M, Lackner K, Rossmann H, et al. Genetics and beyond–the transcriptome of human monocytes and disease susceptibility. PLoS ONE. 2010;5:e10693. doi: 10.1371/journal.pone.0010693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Degner JF, Marioni JC, Pai AA, Pickrell JK, Nkadori E, Gilad Y, Pritchard JK. Effect of read-mapping biases on detecting allele-specific expression from RNA-sequencing data. Bioinformatics. 2009;25:3207–3212. doi: 10.1093/bioinformatics/btp579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kent WJ, Zweig AS, Barber G, Hinrichs AS, Karolchik D. BigWig and BigBed: enabling browsing of large distributed datasets. Bioinformatics. 2010;26:2204–2207. doi: 10.1093/bioinformatics/btq351. [DOI] [PMC free article] [PubMed] [Google Scholar]