Abstract
A vast number of sweet tasting molecules are known, encompassing small compounds, carbohydrates, d-amino acids and large proteins. Carbohydrates play a particularly big role in human diet. The replacement of sugars in food with artificial sweeteners is common and is a general approach to prevent cavities, obesity and associated diseases such as diabetes and hyperlipidemia. Knowledge about the molecular basis of taste may reveal new strategies to overcome diet-induced diseases. In this context, the design of safe, low-calorie sweeteners is particularly important. Here, we provide a comprehensive collection of carbohydrates, artificial sweeteners and other sweet tasting agents like proteins and peptides. Additionally, structural information and properties such as number of calories, therapeutic annotations and a sweetness-index are stored in SuperSweet. Currently, the database consists of more than 8000 sweet molecules. Moreover, the database provides a modeled 3D structure of the sweet taste receptor and binding poses of the small sweet molecules. These binding poses provide hints for the design of new sweeteners. A user-friendly graphical interface allows similarity searching, visualization of docked sweeteners into the receptor etc. A sweetener classification tree and browsing features allow quick requests to be made to the database. The database is freely available at: http://bioinformatics.charite.de/sweet/.
INTRODUCTION
There are three major compounds of life: proteins; lipids and carbohydrates. The perception of sweet taste, mainly associated with advantageous food, has had an important evolutionary influence on different physiological regulation mechanisms. During human development, sugar was always luxury. In 1885 Constantin Fahlberg produced the first artificial sweetener, saccharin, and the scientific establishment was surprised by its extreme sweetness (1). Significant to this discovery was the fact that sweet taste became affordable to poor people. Following the commercial success of artificial sweeteners, a battle between the sugar and sweetener industries began (2). Saccharin was claimed to be carcinogenic in rats (3). However, it was later shown that saccharin is neither toxic nor carcinogenic in normal amounts (4), yet its reputation remains tarnished. Today, the replacement of sugar and other carbohydrates with artificial sweeteners in food is common (5) and is a general approach to prevent cavities (6,7), obesity and associated diseases such as diabetes and hyperlipidemia (8,9).
Currently, the sweet taste receptor, which is a heterodimer of two transmembrane proteins (T1R2 and T1R3) and has several different binding sites, has not been crystallized and is therefore unavailable in the Protein Data Bank (PDB) (10). Such a structure is crucial to elucidating how both small sweeteners and molecules as large as proteins bind and activate the sweet taste receptor (11). In the meantime, modeling studies can provide vital clues to these mechanisms (12). The understanding of compounds binding to the receptor is of relevance not only for the development of new artificial sweeteners but also for improving our understanding of known sweet molecules and what makes them ‘sweet’.
The first publicly available carbohydrate database was CarbBank (13), where users are able to search for carbohydrate structures, sub-structures and non-carbohydrate substituents. Wilhelm von der Lieth established the SweetDB (14), a web-based interface for glycoscientists, which was the basis for further carbohydrate tools collected in the Glycosciences portal (15) and the Glycome-DB (16) that comprises 35 000 carbohydrate sequences with a variety of query options.
There are also a number of databases that deal with glycans. GlycoBase (17) and GlycoEctractor (18) are databases that assist with interpreting high-performance liquid chromatography-glycan profiles. Tyrian Diagnostics used text-mining to develop the GlycoSuiteDB (19), which stores over 7650 glycan structures extracted from 740 papers. The Glycoconjugate Data Bank (20) provides a special tool for N-glycan primary structure verification. The connection to metabolic pathways is provided by KEGG-Glycan (21).
Although, there are a number of resources available with relation to carbohydrates, they are lacking with respect to sweetness and sweeteners. SuperSweet aims to integrate knowledge about the structure of sugars and sweetening agents with receptor binding poses, chemical properties and additional information like sweetness, approval, origin, therapeutic effect and metabolism.
THE DATABASE
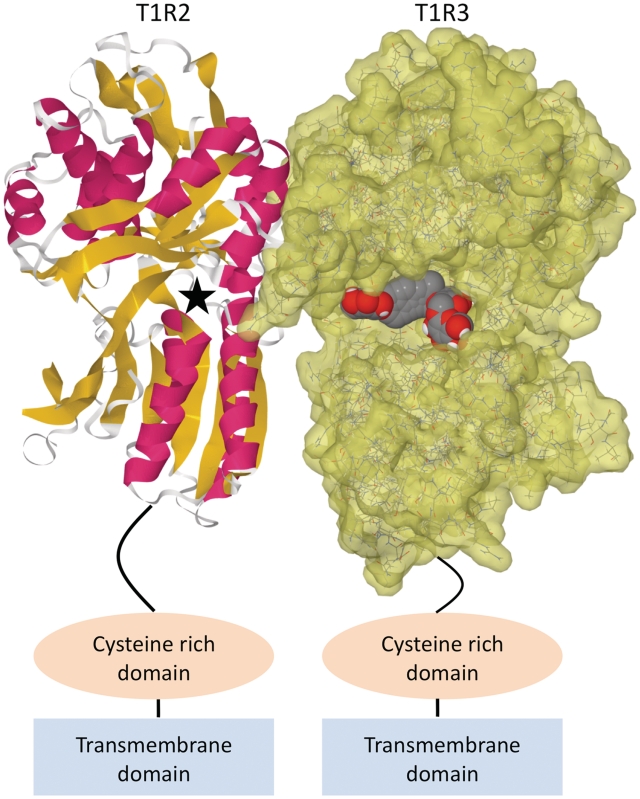
The SuperSweet database was developed for researchers and dieticians and offers a user-friendly interface with helpful examples and FAQs. Currently, SuperSweet comprises more than 8000 carbohydrates, proteins, d-amino acids and artificial (synthesized) sweeteners, which were retrieved from the literature and different pre-existing data sources like Pubchem (22) and the PDB. Similarity searches extended and completed the sweetening agent data set. Besides information about the physicochemical properties of the sweet compounds, the database also offers information about the number of calories, the 3D structure, therapeutic annotations and, if detectable, the sweetness of the molecule. Structural information is available and displayed for each sweet molecule and sweet protein in the database. Moreover, the domain containing the small molecule active site of the sweet receptor was homology modeled and provided in SuperSweet (Figure 1). The small molecules were docked into the modeled binding site and the poses are also stored in the database.
Figure 1.
Homology model of the sweet taste receptor with the sweetening agent Stevioside docked. The T1R2 protomer is displayed in cartoon format and the T1R3 protomer is displayed in wireframe format with a solvent accessible surface rendered (1.2Å probe radius) in transparent yellow. The stevioside molecule is displayed in spacefill format and colored according to atom type. Stevioside was docked into the open protomer, T1R3, as the closed protomer, T1R2, is too small to host large sweeteners. The pocket in the closed protomer is situated on the opposite side to that with stevioside bound—the approximate position is indicated by a black star. The cysteine-rich and transmembrane domains are shown schematically and are not to scale.
There are different options for browsing through the database and for retrieving the data. First, the data can be retrieved by name, physicochemical properties or properties such as calories and sweetness. Secondly, the user is allowed to upload or draw a molecule using the Marvin Sketch plugin (http://www.chemaxon.com). The query structure is compared with the entries of SuperSweet and the results presented in a table comprising molecules and a Tanimoto coefficient expressing their similarity. Thirdly, a sweet-tree is available in SuperSweet that allows the fast and easy selection of a group of sweet tasting molecules (Table 1). Here, the user can find for example, all sweet tasting proteins, or peptides or small molecules like Flavonoids. Finally, SuperSweet offers a browse section, which provides an easy way to access the SuperSweet entries by choosing different categories of molecules based on properties.
Table 1.
Organization of the Sweet-tree
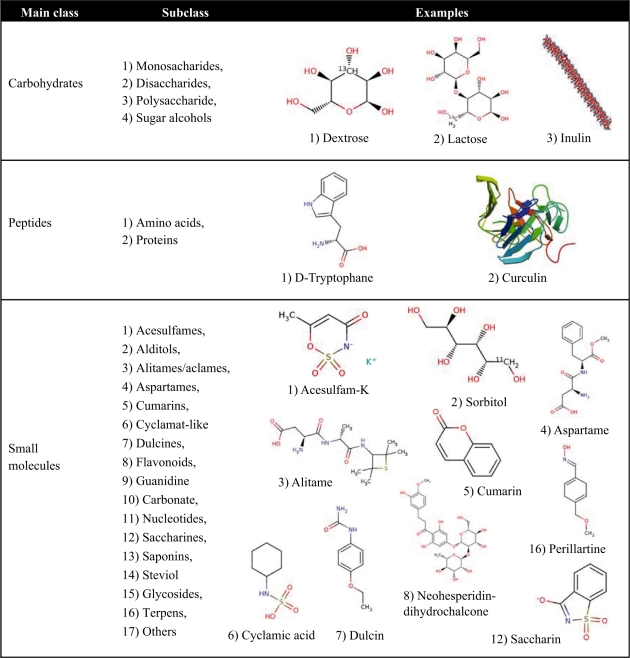 |
The first column presents the three main classes: carbohydrates, peptides and small molecules. The second column shows the subclasses of the main class. The third column shows some examples of the subclasses
MATERIALS AND METHODS
Data acquisition
The sweet tasting molecules were extracted from the literature and publicly available databases like Pubchem, the PDB and MonoSaccharideDB and were filtered using different terms like ‘sweetening agents’. In the next step the data set was extended by using similarity search methods.
Homology modeling of the sweet taste receptor
A model of the large extracellular domain of the sweet taste receptor, which contains two binding sites for small molecular-weight sweeteners, was built using homology modeling. The sweet taste receptor is a class C (or metabotropic) G-protein coupled receptor and exists as a heterodimer (consisting of T1R2 and T1R3) (Figure 1). PSI-BLAST searches (23) of the PDB revealed that T1R2 and T1R3 share ∼25% sequence identity with the metabotropic glutamate receptors and are in close proximity to one another on the phylogentic tree of class C GPCRs (24). As we wanted to use the homology model of the sweet taste receptor for docking studies, it was important that we chose a template structure that is in an active (open-closed) conformation, preferably with a natural ligand bound. Accordingly, an active form of metabotropic glutamate receptor 1 (mGluR1) was used as the template (PDB code: 1EWK) (25), which also had the highest sequence coverage and the highest resolution (2.2Å) compared to other crystal structures of activated glutamate receptors. A multiple sequence alignment was created using MUSCLE (26). The alignment can be downloaded from the SuperSweet website. Homology modeling was carried out using Modeller (27) in Accelrys Discovery Studio 2.5. T1R2 was built using chain A of 1EWK (closed form) and T1R3 using chain B (open form) (12). The large insertions in T1R2 and T1R3 compared to the template were removed from the final model. Lastly, side-chain clashes were removed and the structure was minimized by carrying out 100 steps of both steepest descent and conjugant gradient minimization.
Generation of the binding poses of the small molecules
Docking of the small compounds into the homology modeled receptor was done using the docking program GOLD 4.1.1 (28). In order to define the binding site of the sweet taste receptor, the template structure (mGluR1 containing a glutamate bound to each chain) was superimposed onto the homology model of the sweet taste receptor and the glutamate molecules copied over to the homology model. The binding sites of the sweet taste receptor were then defined by using the glutamate molecules as reference ligands; all atoms within 5Å of the glutamate molecule formed the binding sites for the docking experiments. For each small molecule, 100 docking runs were performed. A previous docking study showed that the sweet taste receptor’s active site in the closed protomer is too small to host some of the larger synthetic sweeteners and is only able to host four compounds out of those tested: saccharin, alitame, aspartame and 6-Cl-tryptophan (12). Experimental work has shown that aspartame and neotame bind to the T1R2 subunit (29). In accordance with these findings, we therefore docked molecules with a molecular weight >400 kDa into T1R3 (open form) and all other molecules into the pockets of both T1R2 and T1R3. The resulting docking poses were then ranked using the GoldScore fitness function. The best scoring docking pose for each molecule can be viewed using a Jmol applet and the respective structure files are also available for download.
Conformer generation
For the small sweetening molecules, conformers were generated using the Accelrys tool. For each small molecule 20 conformers are stored and available for download on the website (30).
Similarity search
For the similarity search in the SuperSweet database, we implemented a bit vector `structural fingerprint’, which encodes the chemical and topological characteristics of a molecule. The fingerprint was pre-calculated for the small molecules of SuperSweet and is also calculated for the query structure, in order to compare it to the database entries. Open Babel implements four different fingerprints (FP2, FP3, FP4 and MACCS). Fingerprint 2 (FP2) is widely used for the comparison of small molecules and is path-based and indexes linear molecules up to seven atoms. However, this fingerprint is not ideal for use in SuperSweet due to its inability to distinguish between different ring structures and therefore between carbohydrates. To overcome this problem, we implemented a combinatorial fingerprint of fingerprint 2 and fingerprint 4. Fingerprint 4 is based on a set of SMARTS patterns and also considers functional groups. For similarity searching the Tanimoto coefficient is used, which gives values in the range of zero (fingerprints have no bits in common) to identical (all bits the same).
Server
SuperSweet is designed as a relational database on a MySQL server. Additionally, the MyChem package (http://mychem.sourceforge.net/) is installed to provide a complete set of functions for handling chemical data within MySQL. Most of the functions used by MyChem depend upon Open Babel (http://openbabel.sourceforge.net/). The structural fingerprint is implemented in Open Babel. To allow the upload or drawing of a query structure, the Marvin Sketch plugin (http://www.chemaxon.com) was installed. For the visualization of the 3D structures Jmol (http://www.jmol.org/) was installed. The website is built with PHP and web access is enabled via Apache HTTP Server 2.2.
EXAMPLE OF USE
Searching for a natural sweetening agent (search field ‘Origin’) with molecular weight between 800 and 900 and sweetness above 200 returns Stevioside. Steviol glycosides like Rebaudioside A or Rubusoside are non-calorific sweeteners that are found, for instance, in sweet Chinese tea (Rubus suavissimus) and Stevia rebaudiana (31). These compounds are of research interest because advantageous effects were observed regarding cancer and blood pressure (32). These effects seem to be the result of binding to other membrane proteins (33). Clicking on the protein icon in the results table shows the docking pose of Stevioside to the sweet receptor (see Figure 1). The structure of Stevioside, the computed conformers, the best docking pose and the modeled receptor structure are downloadable from the website.
More information on Steviol glycosides can be found using the ‘Sweet-tree’ or by performing a search using the field ‘Compound name’ on the ‘Property search’ page. Clicking on the similarity search icon delivers the top 10 similar compounds.
CONCLUSION AND FURTHER DIRECTIONS
SuperSweet compiles information on natural and artificial sweetening agents including their properties such as 3D structure, origin, sweetness, approval, calories etc. and provides hypotheses on their binding to the receptor.
Homology modeling provides a useful means of generating 3D conformations of proteins where experimental structures are not available. For this work, we generated models of the sweet taste receptor using mGluR1. The sequence identity between the receptors is rather low and is within the twilight zone of protein sequence alignments, which makes homology modeling more difficult (34). The quality of our homology model may also be affected by the fact that mGluR1 exists as a homodimer, whereas the sweet taste receptor is a heterodimer. These facts have implications for our docking experiment results due to the strong dependence of docking results on the accuracy of protein structure, especially in the binding site (35). Although GOLD has consistently been shown to be among the best performing docking algorithms in terms of the accuracy of docking poses, it is less able to distinguish the most native-like pose from all of the generated docking poses (36–38). In SuperSweet we have only made the highest scoring pose available for each docked compound and therefore the accuracy of these docking solutions should be considered in light of the aforementioned limitations in homology models and in silico docking.
Unlike the metabotropic glutamate receptors, the sweet taste receptor is predicted to have multiple binding sites (29): (i) two cavities which correspond to the Glu hosting cavities of mGluR1; (ii) a secondary binding site on the surface of the receptor for sweet proteins that corresponds to the wedge model (39) and (iii) overlapping binding sites in the seven transmembrane helix domain for the agonist cyclamate (40) and the inverse agonist lactisole (41). For this work, we only performed docking experiments to the binding sites corresponding to the Glu hosting cavities of mGluR1. These two sites differ in size due to the receptor model being in an open-closed conformation; the binding site in T1R3 is much larger as it is in an open conformation, whereas T1R2 is in a closed conformation. Therefore, we only docked compounds with a molecular weight >400 kDa into T1R3 (open form) and all other compounds into the pockets of both T1R2 and T1R3. In addition, the existence of other binding sites or alternative binding mechanisms cannot be excluded (12). Compared to mGluR1, the additional diversity of compounds binding to the sweet taste receptor and the existence of additional binding sites therefore adds further complexity to in silico docking experiments to the sweet taste receptor.
One of the future goals of SuperSweet is the integration of sugars and sweetening agents into biochemical pathway maps (including PubMed references) to better understand their different ways of metabolism and their impact on metabolic diseases and to foresee possible risk factors. After improving the similarity search and inclusion of pharmacorphore searching to find new putative sweetening agents, a sweetness prediction tool is planned to be implemented. We plan to perform additional text-mining in order to obtain information on the number of calories, sweetness and therapeutic effects of sweet compounds where missing. Docking poses of the sweet proteins to the sweet taste receptor are also planned to be integrated. Another interesting aspect would be the comparison of sweet taste perception with characteristics of sour and bitter taste perception, which is a problem in the development of artificial sweeteners.
AVAILABILITY
The SuperSweet database is freely available under the url: http://bioinformatics.charite.de/sweet/ and will be updated regularly.
FUNDING
Deutsche Forschungsgemeinschaft (SFB 449); Investitionsbank Berlin (IBB); Deutsche Krebshilfe; Bundesministerium für Bildung und Forschung (BMBF); European Union (EU). Funding for open access charge: DFG, BMBF and EU.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank D. Kuzman and A. Chefai for manual curation of the sweetener properties and A.L. Wölke for help with the ‘Sweet tree’.
REFERENCES
- 1.Merki C. Zucker gegen saccharin - zur geschichte der künstlichen süssstoffe. 1993. Campus, Frankfurt/Main/New York. [Google Scholar]
- 2.de la Pena C. Artificial sweetener as a historical window to culturally situated health. Ann. NY Acad. Sci. 2010;1190:159–165. doi: 10.1111/j.1749-6632.2009.05253.x. [DOI] [PubMed] [Google Scholar]
- 3.Hicks RM, Wakefield JS, Chowaniec J. Letter: Co-carcinogenic action of saccharin in the chemical induction of bladder cancer. Nature. 1973;243:347–349. doi: 10.1038/243347a0. [DOI] [PubMed] [Google Scholar]
- 4.Weihrauch MR, Diehl V. Artificial sweeteners—do they bear a carcinogenic risk? Ann. Oncol. 2004;15:1460–1465. doi: 10.1093/annonc/mdh256. [DOI] [PubMed] [Google Scholar]
- 5.Artificial sweeteners: no calories … sweet! FDA Consum. 2006;40:27–28. [PubMed] [Google Scholar]
- 6.Edgar WM. Sugar substitutes, chewing gum and dental caries—a review. Br. Dent. J. 1998;184:29–32. doi: 10.1038/sj.bdj.4809535. [DOI] [PubMed] [Google Scholar]
- 7.Hayes C. The effect of non-cariogenic sweeteners on the prevention of dental caries: a review of the evidence. J. Dent. Educ. 2001;65:1106–1109. [PubMed] [Google Scholar]
- 8.Benton D. Can artificial sweeteners help control body weight and prevent obesity? Nutr. Res. Rev. 2005;18:63–76. doi: 10.1079/NRR200494. [DOI] [PubMed] [Google Scholar]
- 9.Drewnowski A. Intense sweeteners and energy density of foods: implications for weight control. Eur. J. Clin. Nutr. 1999;53:757–763. doi: 10.1038/sj.ejcn.1600879. [DOI] [PubMed] [Google Scholar]
- 10.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Temussi P. The sweet taste receptor: a single receptor with multiple sites and modes of interaction. Adv. Food Nutr. Res. 2007;53:199–239. doi: 10.1016/S1043-4526(07)53006-8. [DOI] [PubMed] [Google Scholar]
- 12.Morini G, Bassoli A, Temussi PA. From small sweeteners to sweet proteins: anatomy of the binding sites of the human T1R2_T1R3 receptor. J. Med. Chem. 2005;48:5520–5529. doi: 10.1021/jm0503345. [DOI] [PubMed] [Google Scholar]
- 13.Doubet S, Albersheim P. CarbBank. Glycobiology. 1992;2:505. doi: 10.1093/glycob/2.6.505. [DOI] [PubMed] [Google Scholar]
- 14.Loss A, Bunsmann P, Bohne A, Loss A, Schwarzer E, Lang E, von der Lieth CW. SWEET-DB: an attempt to create annotated data collections for carbohydrates. Nucleic Acids Res. 2002;30:405–408. doi: 10.1093/nar/30.1.405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lutteke T, Bohne-Lang A, Loss A, Goetz T, Frank M, von der Lieth CW. GLYCOSCIENCES.de: an Internet portal to support glycomics and glycobiology research. Glycobiology. 2006;16:71R–81R. doi: 10.1093/glycob/cwj049. [DOI] [PubMed] [Google Scholar]
- 16.Ranzinger R, Frank M, von der Lieth CW, Herget S. Glycome-DB.org: a portal for querying across the digital world of carbohydrate sequences. Glycobiology. 2009;19:1563–1567. doi: 10.1093/glycob/cwp137. [DOI] [PubMed] [Google Scholar]
- 17.Campbell MP, Royle L, Radcliffe CM, Dwek RA, Rudd PM. GlycoBase and autoGU: tools for HPLC-based glycan analysis. Bioinformatics. 2008;24:1214–1216. doi: 10.1093/bioinformatics/btn090. [DOI] [PubMed] [Google Scholar]
- 18.Artemenko NV, Campbell MP, Rudd PM. GlycoExtractor: a web-based interface for high throughput processing of HPLC-glycan data. J. Proteome Res. 2010;9:2037–2041. doi: 10.1021/pr901213u. [DOI] [PubMed] [Google Scholar]
- 19.Cooper CA, Joshi HJ, Harrison MJ, Wilkins MR, Packer NH. GlycoSuiteDB: a curated relational database of glycoprotein glycan structures and their biological sources 2003 update. Nucleic Acids Res. 2003;31:511–513. doi: 10.1093/nar/gkg099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Nakahara T, Hashimoto R, Nakagawa H, Monde K, Miura N, Nishimura S. Glycoconjugate Data Bank: Structures—an annotated glycan structure database and N-glycan primary structure verification service. Nucleic Acids Res. 2008;36:D368–D371. doi: 10.1093/nar/gkm833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hashimoto K, Kawano S, Goto S, Aoki-Kinoshita KF, Kawashima M, Kanehisa M. A global representation of the carbohydrate structures: a tool for the analysis of glycan. Genome Inform. 2005;16:214–222. [PubMed] [Google Scholar]
- 22.Wang Y, Xiao J, Suzek TO, Zhang J, Wang J, Bryant SH. PubChem: a public information system for analyzing bioactivities of small molecules. Nucleic Acids Res. 2009;37:W623–W633. doi: 10.1093/nar/gkp456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Altschul SF, Madden TL, Schaffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fredriksson R, Schioth HB. The repertoire of G-protein-coupled receptors in fully sequenced genomes. Mol. Pharmacol. 2005;67:1414–1425. doi: 10.1124/mol.104.009001. [DOI] [PubMed] [Google Scholar]
- 25.Kunishima N, Shimada Y, Tsuji Y, Sato T, Yamamoto M, Kumasaka T, Nakanishi S, Jingami H, Morikawa K. Structural basis of glutamate recognition by a dimeric metabotropic glutamate receptor. Nature. 2000;407:971–977. doi: 10.1038/35039564. [DOI] [PubMed] [Google Scholar]
- 26.Edgar RC. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res. 2004;32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sali A, Blundell TL. Comparative protein modelling by satisfaction of spatial restraints. J. Mol. Biol. 1993;234:779–815. doi: 10.1006/jmbi.1993.1626. [DOI] [PubMed] [Google Scholar]
- 28.Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein-ligand docking using GOLD. Proteins. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- 29.Xu H, Staszewski L, Tang H, Adler E, Zoller M, Li X. Different functional roles of T1R subunits in the heteromeric taste receptors. Proc. Natl Acad. Sci. USA. 2004;101:14258–14263. doi: 10.1073/pnas.0404384101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Gunther S, Senger C, Michalsky E, Goede A, Preissner R. Representation of target-bound drugs by computed conformers: implications for conformational libraries. BMC Bioinformatics. 2006;7:293. doi: 10.1186/1471-2105-7-293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Geuns JM. Stevioside. Phytochemistry. 2003;64:913–921. doi: 10.1016/s0031-9422(03)00426-6. [DOI] [PubMed] [Google Scholar]
- 32.Takasaki M, Konoshima T, Kozuka M, Tokuda H, Takayasu J, Nishino H, Miyakoshi M, Mizutani K, Lee KH. Cancer preventive agents. Part 8: chemopreventive effects of stevioside and related compounds. Bioorg. Med. Chem. 2009;17:600–605. doi: 10.1016/j.bmc.2008.11.077. [DOI] [PubMed] [Google Scholar]
- 33.Chatsudthipong V, Muanprasat C. Stevioside and related compounds: therapeutic benefits beyond sweetness. Pharmacol. Ther. 2009;121:41–54. doi: 10.1016/j.pharmthera.2008.09.007. [DOI] [PubMed] [Google Scholar]
- 34.Rost B. Twilight zone of protein sequence alignments. Protein Eng. 1999;12:85–94. doi: 10.1093/protein/12.2.85. [DOI] [PubMed] [Google Scholar]
- 35.Bordogna A, Pandini A, Bonati L. Predicting the accuracy of protein-ligand docking on homology models. J. Comput. Chem. doi: 10.1002/jcc.21601. [Epub ahead of print, 6 July 2010]; PMID: 20607693. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kellenberger E, Rodrigo J, Muller P, Rognan D. Comparative evaluation of eight docking tools for docking and virtual screening accuracy. Proteins. 2004;57:225–242. doi: 10.1002/prot.20149. [DOI] [PubMed] [Google Scholar]
- 37.Kontoyianni M, McClellan LM, Sokol GS. Evaluation of docking performance: comparative data on docking algorithms. J. Med. Chem. 2004;47:558–565. doi: 10.1021/jm0302997. [DOI] [PubMed] [Google Scholar]
- 38.Plewczynski D, Lazniewski M, Augustyniak R, Ginalski K. Can we trust docking results? Evaluation of seven commonly used programs on PDBbind database. J. Comput. Chem. doi: 10.1002/jcc.21643. [Epub ahead of print, 1 September 2010]; PMID: 20812323. [DOI] [PubMed] [Google Scholar]
- 39.Temussi PA. Why are sweet proteins sweet? Interaction of brazzein, monellin and thaumatin with the T1R2-T1R3 receptor. FEBS Lett. 2002;526:1–4. doi: 10.1016/s0014-5793(02)03155-1. [DOI] [PubMed] [Google Scholar]
- 40.Jiang P, Cui M, Zhao B, Snyder LA, Benard LM, Osman R, Max M, Margolskee RF. Identification of the cyclamate interaction site within the transmembrane domain of the human sweet taste receptor subunit T1R3. J. Biol. Chem. 2005;280:34296–34305. doi: 10.1074/jbc.M505255200. [DOI] [PubMed] [Google Scholar]
- 41.Jiang P, Cui M, Zhao B, Liu Z, Snyder LA, Benard LM, Osman R, Margolskee RF, Max M. Lactisole interacts with the transmembrane domains of human T1R3 to inhibit sweet taste. J. Biol. Chem. 2005;280:15238–15246. doi: 10.1074/jbc.M414287200. [DOI] [PubMed] [Google Scholar]