Abstract
COSMIC (http://www.sanger.ac.uk/cosmic) curates comprehensive information on somatic mutations in human cancer. Release v48 (July 2010) describes over 136 000 coding mutations in almost 542 000 tumour samples; of the 18 490 genes documented, 4803 (26%) have one or more mutations. Full scientific literature curations are available on 83 major cancer genes and 49 fusion gene pairs (19 new cancer genes and 30 new fusion pairs this year) and this number is continually increasing. Key amongst these is TP53, now available through a collaboration with the IARC p53 database. In addition to data from the Cancer Genome Project (CGP) at the Sanger Institute, UK, and The Cancer Genome Atlas project (TCGA), large systematic screens are also now curated. Major website upgrades now make these data much more mineable, with many new selection filters and graphics. A Biomart is now available allowing more automated data mining and integration with other biological databases. Annotation of genomic features has become a significant focus; COSMIC has begun curating full-genome resequencing experiments, developing new web pages, export formats and graphics styles. With all genomic information recently updated to GRCh37, COSMIC integrates many diverse types of mutation information and is making much closer links with Ensembl and other data resources.
INTRODUCTION
COSMIC is designed to gather, curate, organize and present the world’s information on somatic mutations in cancer and make it freely available in a variety of useful ways, most easily accessible through its website (http://www.sanger.ac.uk/cosmic). As previously described (1,2), COSMIC combines cancer mutation data manually curated from the scientific literature, with the output from the Cancer Genome Project (CGP) at the Sanger Institute UK. Genes are selected for full literature curation using the Cancer Gene Census (http://www.sanger.ac.uk/genetics/CGP/Census/), with a focus on those mutated by small point mutations in the coding domains, and more recently including those mutated by gene fusion. In the age of whole cancer genome sequencing, it is now possible to describe the genome-wide somatic mutation content of a tumour sample, including structural rearrangements and non-coding variants. COSMIC is now integrating this information into the database, providing full coding and genomic variant annotations for samples, both from CGP laboratories and recent publications.
DATABASE CONTENT
For years, the COSMIC database has described somatic mutations in key cancer genes across many cancer samples, and, more recently, gene fusions and structural rearrangement annotations have been included. Much of the basic functionality of the system has been maintained as previously described (1,2), including the nomenclature system for tumour phenotypes and the HGVS syntax for mutations. In the past year, full mutation details have been curated into COSMIC for 19 new cancer genes, making a total of 83 fully curated and up-to-date cancer genes, the majority of which are point-mutated; the number of curated fusion gene pairs has also doubled to 49. In addition to this, a major effort to curate significant external resources and publications into COSMIC has enhanced the coverage of the database. Most importantly, the mutation spectrum of the key cancer gene TP53 is now included; in collaboration with the curators at IARC (3), the majority of the IARC p53 database R14 release is now available in COSMIC. In addition, all the somatic mutation annotations from the TCGA large-scale cancer characterization project are now included (4), as are those from a number of significant systematic candidate gene screen publications (5–7). In total, more than 47 000 coding mutation annotations have been added to COSMIC in the last year, together with over 53 000 non-coding mutations. Current contents of the database (v48, July 2010) are displayed in Table 1.
Table 1.
Total contents in v48 of the COSMIC database, July 2010 release
| Curated data type | Curated data count |
|---|---|
| Experiments | 2 760 220 |
| Tumours | 541 928 |
| Mutations | 136 326 |
| References | 10 383 |
| Genes | 18 490 |
| Fusions | 4946 |
| Structural variants | 2307 |
| Whole cancer genomes | 29 |
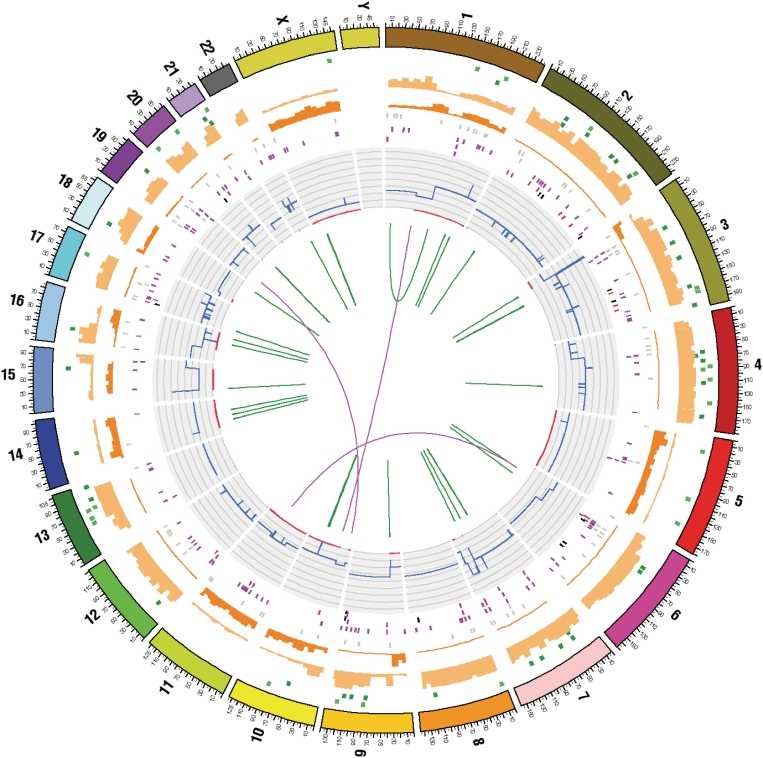
With the increasing genomic context in COSMIC, the system has expanded to encompass the complete annotation of somatic mutations across whole genomes, including all coding and non-coding mutations, structural rearrangements and gene fusions. Links are also present into the CONAN system (http://www.sanger.ac.uk/cgi-bin/genetics/CGP/cghviewer/CghHome.cgi) for copy number variant annotations. The fullest and most detailed genome annotations are from the CGP laboratories (8,9), but in addition, curation of large-scale systematic candidate gene screens (5,6,7) and whole genome analyses (10) from the scientific literature has commenced. Twenty-nine tumour samples now have genome-wide annotations in COSMIC. The first 26 samples, mostly breast carcinoma tumours, represent low-coverage paired-end scans characterizing somatic structural rearrangements at basepair resolution (11). Two samples (malignant melanoma and small-cell lung carcinoma cell lines) have undergone whole-genome resequencing, providing much more extensive annotations (8,9). Point mutations and small insertions/deletions are described, several hundred in coding domains and many thousands either intronic or intergenic. Including structural variants and CNVs, these samples are summarized in a more complex circos diagram, presenting rings describing point mutation types and impact on any coding domains (Figure 1). Finally, Mardis et al. (10) describe the full-genome resequencing of a single AML sample (and subsequent characterization of a further cohort); this is the first genome analysis to be curated from the literature. All non-coding variants and structural rearrangements are described only in terms of GRCh37 co-ordinates, whilst coding mutations are described in genomic terms, together with co-ordinates in the CDS and peptide they affect.
Figure 1.
Circos diagram summarizing the full somatic mutation content of cell line NCI-H209. Concentric rings summarize the data on different types of mutation. From the inside out, the core displays the structural rearrangements; intrachromosomal are in green, interchromosomal in purple. The next ring out shows the chromosomal copy number in histogram form, with inner red patches indicating regions of LOH. Further out, several rings of single base coding substitutions are shown (black tiles show splice site mutations, red stop-gained, purple non-synonymous and grey synonymous changes). The inner dark orange and outer light orange histograms represent non-coding mutations, relative frequencies of homozygous and heterozygous mutations, respectively. In the final ring before the chromosome indicators, indels are shown in green; light green represents insertions and dark green deletions.
DATA ACCESS
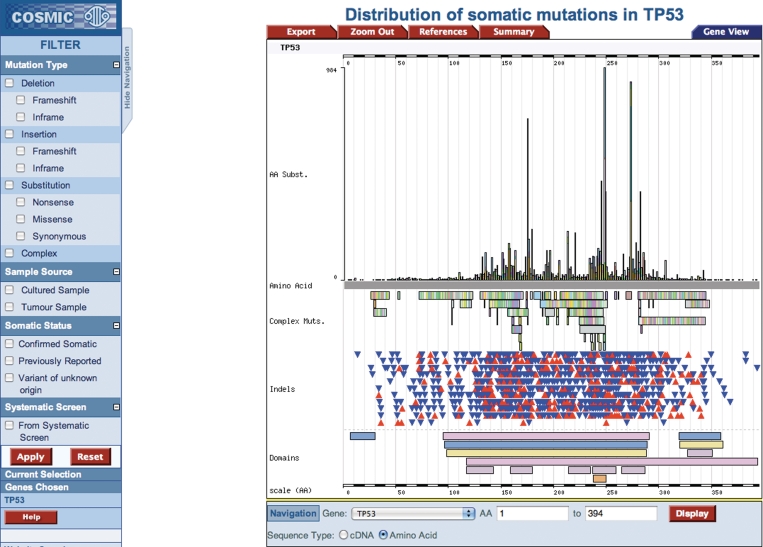
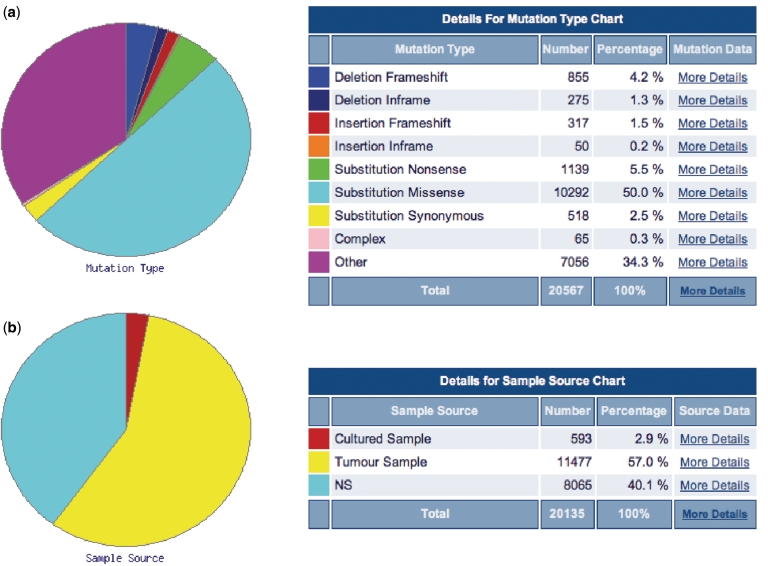
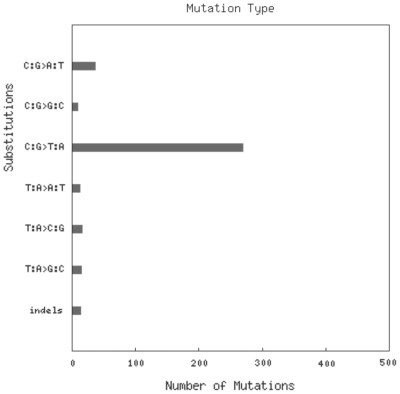
The COSMIC website is available at http://www.sanger.ac.uk/cosmic/. While the ability to navigate the COSMIC database by gene and tissue type has been maintained, it has evolved to become much more mineable. The gene histogram page, which graphically summarizes the somatic mutations on the coding sequence of the gene (e.g. Figure 2), still forms the core of the navigation system for the majority of the data. Multiple methods are now available for filtering the data, forming specialized queries. In each case, the image will be regenerated, and the mutation spectrum and tissue-specific frequencies will be recalculated for the selection chosen: (i) clicking on the graphic or inputting CDS co-ordinates zooms into the required region of the gene; (ii) clicking on the primary tissue type offers a method of selecting a specialized phenotype; and (iii) in the left-side navigation bar (left side of Figure 2), further filters are available including restrictions by mutation type (substitution, deletion, etc.; missense, nonsense, frameshift etc.), sample source (cell line, primary tumour), somatic status (was the mutation confirmed somatic or was the normal tissue not available) and systematic screen (were these results generated as part of a much larger genome-wide candidate gene screen or genome resequencing study). Simultaneously using multiple filters, it is possible to build up a very specific query focused on exact requirements of gene/phenotype and data content. Expanding this selection process, we have begun generating summary diagrams in the form of pie charts which both overview the selected data and provide links into subsets by automating the selection process in one click. Initially available under the ‘Distribution’ button on the histogram page are two such summaries (Figure 3): the first provides a breakdown of mutation counts by mutation type and the second shows a breakdown of mutant samples by their source (cell line, primary tumour or unknown). In each case, ‘More Details’ links provide options to regenerate the histogram page with the specified selection, or to view the full data in tabulated form, ready for export in spreadsheet format. In addition to enabling mutation spectrum analyses at the gene level, COSMIC has also begun providing spectrum analyses at the sample level. For samples with significant numbers of mutation data, a mutation spectrum histogram is available (Figure 4) to show summary nucleotide exchange frequencies from the sample’s repertoire of coding mutations. This can be found on the sample overview page, where multiple tabs facilitate the examination of many data types for each sample.
Figure 2.
The gene histogram page for TP53. The histogram shows relative frequencies of mutations (y-axis) across the CDS of the gene (x-axis). Underneath the x-axis scale bar are complex replacement mutations, followed by simple deletions (blue triangles) and insertions (red triangles). Under this, zoom options are available. On the left, the new specialization filters are shown, offering many query options.
Figure 3.
Pie charts (here showing the TP53 gene) are increasingly used for summarization of complex spectrum data in COSMIC. Two are currently live with many more forthcoming. The top graph (a) shows the breakdown of all observed mutations by type, and the lower (b) shows the breakdown of mutated samples by source. The total number differs slightly due to some samples having more than one mutation, thus being counted once in (b) but twice or more in (a).
Figure 4.
Mutation spectrum histogram for whole-genome-resequencing sample COLO-829, displaying the considerable overrepresentation of C:G>T:A events in its coding mutation repertoire, reflecting the characteristic signature of DNA damage due to ultraviolet light exposure common in malignant melanoma.
While the website has been built to be as user friendly as possible, a new Biomart (12) has been made available which emphasizes flexibility (available at http://www.sanger.ac.uk/genetics/CGP/cosmic/biomart/martview/). This system provides all available selections on genes, tissues and mutations as pull-down menus and provides tabulated reports on the data selected which are again exportable in spreadsheet format for offline investigation.
Data integration and interoperability
An increasing emphasis is developing on resequencing of full cancer genomes and, in response, we have upgraded all our genomic co-ordinates to GRCh37. This has allowed us to begin integrating much more directly with the Ensembl genome browser (www.ensembl.org). Data on the 83 curated genes have been uploaded from COSMIC into Ensembl databases, and this has allowed the incorporation of COSMIC data directly into the Ensembl web pages, as ‘Somatic_SNV’ annotations (distinguishing the somatic cancer mutations from standard SNPs). These pages display the COSMIC mutation in the local sequence context, with annotations on all Ensembl transcripts for the gene. Also included are links to Ensembl’s GenomeView websystem, providing views of the mutated gene with full genome annotations. Links back to COSMIC have been carefully incorporated into the Ensembl website to give more detailed and specialist views of the data. Initially released in August 2010, the full COSMIC data set is expected shortly after.
A number of other external projects have been receiving our support integrating COSMIC data, including Intogen (http://www.intogen.org/home), UniProt (http://www.ebi.ac.uk/uniprot) and Pfam (http://pfam.sanger.ac.uk). Many more have used exports from the FTP site to extend existing systems (e.g. CGWB; https://cgwb.nci.nih.gov), or develop new ones, for example, as integration resources of selected data subsets (e.g. CanProVar; http://bioinfo.vanderbilt.edu/canprovar). Further external projects have integrated COSMIC’s search feature into their systems, interpreting results of remote search queries for local examination, with links back into the COSMIC website (e.g. ONIX; http://www.ncri-onix.org.uk). A stable identifier system has been developed for COSMIC somatic mutations to allow external databases to easily link back to the appropriate COSMIC record. All mutations held in COSMIC are assigned a ‘COSM’ id (COsmic Somatic Mutation identifier) which will remain stable between releases of COSMIC. Ensembl is the first external database to successfully use the COSM id and we ask other database to maintain this identifier when using COSMIC annotated mutations.
FUTURE WORK
The data in COSMIC are continually updated, to maintain the existing curated genes and to include new fully curated genes—this work will continue. Increasing numbers of genes are also being added during the curation of large-scale candidate gene screens which can encompass more than 20 000 genes in one study. More significantly, there are increasingly numerous studies detailing the full resequencing of entire cancer genomes; while the first few have already been released in COSMIC, it is expected that this will prove a major focus for COSMIC’s development. While genome data have already been exported from COSMIC into the ICGC (www.icgc.org), it is intended that COSMIC should also import ICGC-validated somatic mutation data to aggregate with genomes curated from elsewhere, maximizing the cancer genome coverage of the COSMIC database.
In order to effectively interrogate this increasingly huge amount of information, new tools are being developed which provide new functionality currently unavailable in COSMIC. Already scheduled for a late 2010 release is a GBrowse system embedded within COSMIC. GBrowse (13) is a fully featured and very flexible genome browser, and, to work within COSMIC, it has been populated with full genome annotations alongside which COSMIC data are easily navigable in a genomic context. It combines into one window most of the data currently available in COSMIC, including all gene structures and sequences, all point mutations, structural rearrangements and copy number aberrations. In addition to improving the genomic context of COSMIC, a new analytical suite is being built for the main gene-centric system also. In similar ways to the existing mutation-type pie charts (Figure 3), new charts and tables are being designed to display mutation information according to constraints such as basepair sequence change or insertion/deletion size. These will be especially powerful, as they will employ all the specialization filters that are used in deeply examining the gene histogram page as described earlier. The COSMIC project has been running for over 9 years, and is supported to continue for many more. A stable and comprehensive resource, it is now meeting the challenge of annotating and integrating a wide range of somatic mutation data from many new sources, continuing to make it easily and freely available to the research community.
FUNDING
The Wellcome Trust supported this work under grant reference 077012/Z/05/Z. Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to thank Magali Olivier for her substantial help interpreting the IARC p53 R14 database for upload into COSMIC.
REFERENCES
- 1.Forbes SA, Tang G, Bindal N, Bamford S, Dawson E, Cole C, Kok CY, Jia M, Ewing R, Menzies A, et al. COSMIC (the Catalogue of Somatic Mutations in Cancer): a resource to investigate acquired mutations in human cancer. Nucleic Acids Res. 2010;38:D652–D657. doi: 10.1093/nar/gkp995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Forbes SA, Bhamra G, Bamford S, Dawson E, Kok C, Clements J, Menzies A, Teague JW, Futreal PA, Stratton MR. The Catalogue of Somatic Mutations in Cancer (COSMIC) Curr. Protoc. Hum. Genet. 2008 doi: 10.1002/0471142905.hg1011s57. Chapter 10, 11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Petitjean A, Mathe E, Kato S, Ishioka C, Tavtigian SV, Hainaut P, Olivier M. Impact of mutant p53 functional properties on TP53 mutation patterns and tumor phenotype: lessons from recent developments in the IARC TP53 database. Hum. Mutat. 2007;28:622–629. doi: 10.1002/humu.20495. [DOI] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Comprehensive genomic characterization defines human glioblastoma genes and core pathways. Nature. 2008;455:1061–1068. doi: 10.1038/nature07385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sjöblom T, Jones S, Wood LD, Parsons DW, Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al. The consensus coding sequences of human breast and colorectal cancers. Science. 2006;314:268–274. doi: 10.1126/science.1133427. [DOI] [PubMed] [Google Scholar]
- 6.Parsons DW, Jones S, Zhang X, Lin JC, Leary RJ, Angenendt P, Mankoo P, Carter H, Siu IM, Gallia GL, et al. An integrated genomic analysis of human glioblastoma multiforme. Science. 2008;321:1807–1812. doi: 10.1126/science.1164382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Ding L, Getz G, Wheeler DA, Mardis ER, McLellan MD, Cibulskis K, Sougnez C, Greulich H, Muzny DM, Morgan MB, et al. Somatic mutations affect key pathways in lung adenocarcinoma. Nature. 2008;455:1069–1075. doi: 10.1038/nature07423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pleasance ED, Stephens PJ, O'Meara S, McBride DJ, Meynert A, Jones D, Lin ML, Beare D, Lau KW, Greenman C, et al. A small-cell lung cancer genome with complex signatures of tobacco exposure. Nature. 2010;463:184–190. doi: 10.1038/nature08629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pleasance ED, Cheetham RK, Stephens PJ, McBride DJ, Humphray SJ, Greenman CD, Varela I, Lin ML, Ordóñez GR, Bignell GR, et al. A comprehensive catalogue of somatic mutations from a human cancer genome. Nature. 2009;463:191–196. doi: 10.1038/nature08658. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mardis ER, Ding L, Dooling DJ, Larson DE, McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath SD, et al. Recurring mutations found by sequencing an acute myeloid leukemia genome. N. Engl. J. Med. 2009;361:1058–1066. doi: 10.1056/NEJMoa0903840. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Stephens PJ, McBride DJ, Lin ML, Varela I, Pleasance ED, Simpson JT, Stebbings LA, Leroy C, Edkins S, Mudie LJ, et al. Complex landscapes of somatic rearrangement in human breast cancer genomes. Nature. 2009;462:1005–1010. doi: 10.1038/nature08645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart central portal – unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]