Abstract
The concept of homology drives speculation on a gene’s function in any given species when its biological roles in other species are characterized. With reference to a specific species radiation homologous relations define orthologs, i.e. descendants from a single gene of the ancestor. The large-scale delineation of gene genealogies is a challenging task, and the numerous approaches to the problem reflect the importance of the concept of orthology as a cornerstone for comparative studies. Here, we present the updated OrthoDB catalog of eukaryotic orthologs delineated at each radiation of the species phylogeny in an explicitly hierarchical manner of over 100 species of vertebrates, arthropods and fungi (including the metazoa level). New database features include functional annotations, and quantification of evolutionary divergence and relations among orthologous groups. The interface features extended phyletic profile querying and enhanced text-based searches. The ever-increasing sampling of sequenced eukaryotic genomes brings a clearer account of the majority of gene genealogies that will facilitate informed hypotheses of gene function in newly sequenced genomes. Furthermore, uniform analysis across lineages as different as vertebrates, arthropods and fungi with divergence levels varying from several to hundreds of millions of years will provide essential data for uncovering and quantifying long-term trends of gene evolution. OrthoDB is freely accessible from http://cegg.unige.ch/orthodb.
INTRODUCTION
Recognizing similarities as evidence of shared ancestry describes the general biological concept of homology that can be applied specifically to genes encoded in complete genomes to delineate orthologs, the ‘equivalent’ genes in different species, and paralogs, gene duplicates within one genome (1–3). The rapidly increasing number of sequenced genomes presents a remarkable opportunity as well as a formidable challenge to resolve complex gene histories in unprecedented detail. As orthologous relations are defined by speciation events where orthologs arise by vertical decent from a single gene of the last common ancestor, such classifications are inherently hierarchical. Gene duplication events after speciation disrupt the 1:1 correspondence of genes among species and lead to the formation of orthologous groups, comprised of all genes descended from a single gene of the last common ancestor. Many algorithms have been developed to apply these principles and meet the challenge of large-scale data analysis (4,5). These can be broadly classified into those that cluster the results from all-against-all pairwise sequence comparisons and those that employ phylogenetic tree-based methods. The growing number of different resources for cluster-based approaches [e.g. (6–12)], as well as for phylogenetic approaches [e.g. (13–20)], reflects the importance of ortholog identification as a cornerstone of comparative genomics that drives evolutionary and molecular biology research.
The preservation of orthologs across many species over long evolutionary periods, especially as single-copy genes, strongly supports hypotheses of conserved functionality (21). By contrast, duplication events may allow for functional divergence (22). Thus, although orthologous relations are not defined by gene function, inferences of common functions remain the most plausible evolutionary scenario and therefore justify one of the major objectives of orthology delineation: the tentative transfer of functional annotations from well-studied organisms to the newly sequenced species (23).
Here, we present the update of OrthoDB (10), the hierarchical catalog of eukaryotic orthologs, featuring an expanded species sampling, extensive functional and evolutionary annotation of the derived orthologous groups, as well as improved text-based searches, querying on the phyletic profile of orthologous gene copy numbers, and searches by sequence homology. OrthoDB is freely accessible from http://cegg.unige.ch/orthodb, and now referenced with link-outs from a number of resources including UniProt (24) and FlyBase (25).
METHODOLOGY
Orthology is defined relative to the last common ancestor of the species being considered, thereby determining the hierarchical nature of orthologous classifications (1–3) (Supplementary Figure S1). This is explicitly addressed in OrthoDB (10) by application of the orthology delineation procedure at each radiation point of the considered phylogeny, empirically computed over the super-alignment of single-copy orthologs using a maximum-likelihood approach corroborated with known taxonomies. The OrthoDB implementation employs a BRH clustering algorithm based on all-against-all Smith–Waterman (26) protein sequence comparisons computed using PARALIGN (27). Gene set pre-processing selects the longest protein-coding transcript of alternatively spliced genes and of very similar gene copies (>97% identity). The newly optimized procedure triangulates BRHs with an e-value cutoff of 1e-3 to progressively build the clusters, (non-triangulated BRHs are considered with an e-value cutoff of 1e-6), and requiring an overall minimum sequence alignment overlap of 30 amino acids to avoid domain walking. These core clusters are further expanded to include all more closely related within-species in-paralogs, and the previously identified very similar gene copies. Inspections of the OrthoDB orthologous classifications as part of several genome projects (28–31) and other comparative genomic studies (32–35) have confirmed their biological relevance and acceptable accuracy.
UPDATED DATABASE CONTENT
Gene sets
The complete predicted protein-coding gene sets were retrieved from publically available genomic resources including 44 vertebrates from Ensembl (36) (Release 58, May 2010), 25 arthropods from AphidBase (37), BeetleBase (38), FlyBase (25), Hymenoptera Genome Database, SilkDB (39), VectorBase (40) and wFleaBase (41) (current releases in July 2010), and 46 fungi from UniProt (24) (August 2010 release). Gene sets for an additional five animal species were retrieved for orthology delineation across metazoa: lancelet, polyp, sea anemone, sea urchin, and worm (current releases in July 2010). For full details of the genome assembly and gene set releases used for each species, please see Supplementary Table S1.
Gene annotations
Annotations describing putative functional attributes were sourced from UniProt (24), as well as from species-specific resources including Mouse Genome Informatics (MGI) (42), FlyBase (25) and Saccharomyces Genome Database (SGD) (43). UniProt identifier cross-referencing allowed mapping of gene annotations to the gene sets retrieved from Ensembl and other sources. The UniProt data were also employed to comprehensively map gene names and synonyms, as well as secondary gene identifiers and cross-referenced database gene identifiers, e.g. RefSeq, Entrez GeneID, GenBank, Protein Data Bank and Mendelian Inheritance in Man, as well as assigned Gene Ontology (GO) (44) attributes. The species-specific model organism databases (MGI, FlyBase and SGD) provided mapping to additional gene synonyms and identifiers as well as selected controlled-vocabulary gene phenotypes from relevant experimental data (Supplementary Table S2). Protein domain signatures were retrieved from InterPro (45) matches to the UniProt Archive (UniParc) of non-redundant protein sequences.
Orthologous groups
Analysis of the selected eukaryotic species focused on resolving orthologous relations at each radiation of the three sampled lineages, as well as delineating metazoan orthologous groups by analyzing the vertebrates and arthropods with five additional animal species. For the complete sets of vertebrates, arthropods and fungi, 87% of a total of 1 611 843 genes were classified into 18 474, 20 428 and 14 088 orthologous groups, respectively (Table 1). The greater spans and faster evolutionary rates across the arthropod and fungal phylogenies (33,46,47) may limit the detection of very distant homology, leading to the observed lower proportions of classified genes compared to the vertebrates. Additional factors that may influence the proportions of classified genes include the completeness and coverage of genome sequencing as well as quality and consistency of gene repertoire predictions (e.g. variable strategies applied to arthropods).
Table 1.
OrthoDB species and gene content
| Lineage | Species count | Input genes |
Classified genesa |
Orthologous groupsa | |||
|---|---|---|---|---|---|---|---|
| Total | Average | SD | Count | % | |||
| Vertebrates | 44 | 793 077 | 18 024 | 2884 | 748 195 | 94 | 18 474 |
| Arthropods | 25 | 422 677 | 16 907 | 5280 | 325 685 | 77 | 20 428 |
| Fungi | 46 | 396 089 | 8611 | 3632 | 331 623 | 84 | 14 088 |
Statistics describing the input gene sets and OrthoDB classifications.
aTotal counts of genes and orthologous groups at the root of each lineage.
NEW OrthoDB FEATURES
Functional annotations of orthologous groups
Orthology delineation aims to identify groups of genes descended from a common ancestor, thereby enabling tentative functional attributes ascribed to one or more members to be generally extrapolated to describe the group as a whole. Protein-coding genes from model organisms are by far the best studied and therefore provide the most comprehensive annotations and insights into biological functions; however, ill-informed extrapolation of annotation across species can lead to error propagation. Leaving this to the expert, we merely summarize the available functional evidence of orthologous genes that is indicative of their common functional role.
GO and protein domain summaries
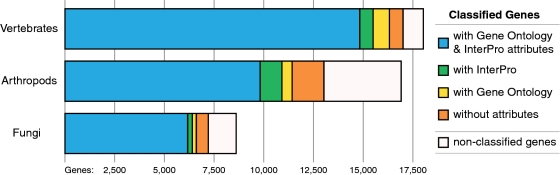
OrthoDB orthologous group functional annotations are summarized from associated GO and InterPro attributes of individual genes, supplemented by data from representative model organisms. Of the just over 1.4 million orthologous group member genes, almost 95% are classified in orthologous groups that can be described by either GO terms (molecular function, biological process or cellular component) or InterPro domains, and more than 85% by both attributes (Figure 1, Supplementary Figure S2). For each orthologous group, summarizing the member gene GO (molecular function, biological process and cellular component) and InterPro annotations highlights the functional attributes that describe the orthologous group as a whole (Figure 2). These descriptions identify the frequencies of associated GO terms together with InterPro domains of member genes, and list succinct term and domain descriptions.
Figure 1.
Striving for the most complete coverage with acceptable accuracy, OrthoDB provides tentative associations of the vast majority of classified genes with Gene Ontology (GO) and InterPro functional attributes.
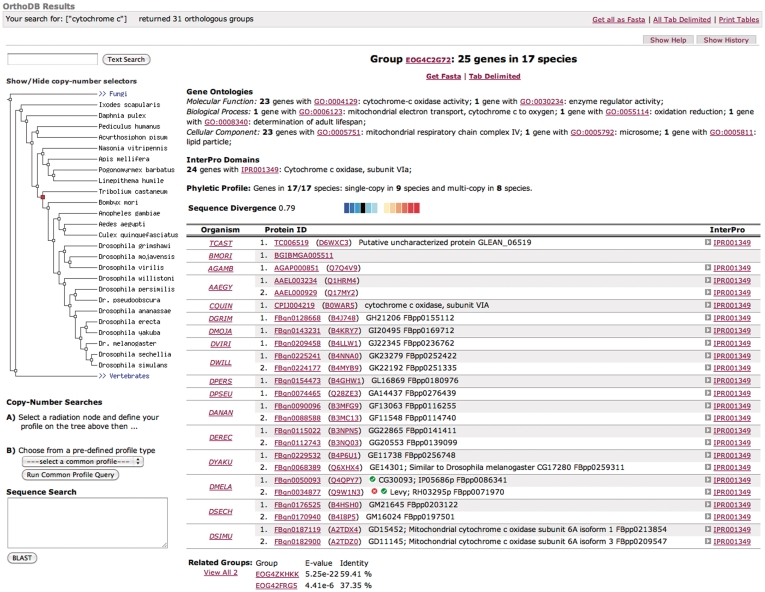
Figure 2.
Screenshot of a sample result page, featuring functional Gene Ontology (GO), InterPro and phenotype annotations, as well as evolutionary related groups, phyletic profile and relative divergence among orthologs.
Model organism phenotypes
Mapping of selected model organism phenotype data identifies a significant proportion of orthologous groups with genes from model organisms that exhibit experimental phenotypes. This approach therefore facilitates querying of OrthoDB with key function-related terms from the respective phenotype ontologies, e.g. sterile, or cell cycle defective (Supplementary Table S2). For the representative model organisms in each lineage (Mus musculus, Drosophila melanogaster or Saccharomyces cerevisiae), gene synonyms and secondary identifiers, as well as selected associated phenotypes, are indicated with distinct icons linked to their respective database sources.
In addition, for each orthologous group member gene, concise UniProt functional descriptors are provided with links to the mapped entries. InterPro matches are displayed with domains ordered sequentially from the N- to C-terminus, describing the complete domain architecture of multidomain genes. The orthologous group summary annotations together with the attributes of individual gene members provide a snapshot of the available functional information, with extensive links to respective source databases, allowing further investigation of their putative biological roles.
Evolutionary annotations of orthologous groups
Protein sequence divergence rate among orthologous group member genes, their phyletic gene copy-number profiles and their homology to genes in other orthologous groups are indicators of the level of confidence with which functional annotations from genes of well-studied model organisms may be transferred to other species. Evolutionary annotations of orthologous groups therefore complement the functional annotations by presenting these quantifiable evolutionary properties (Figure 2).
Evolutionary rates
Orthologous groups that exhibit appreciably higher or lower levels of sequence divergence are highlighted through quantification of the relative divergence among their member genes. These are computed for each orthologous group as the average of interspecies identities normalized to the average identity of all interspecies BRHs, computed from pairwise Smith–Waterman alignments of protein sequences (Supplementary Figure S3).
Phyletic profiles
Orthologous group phyletic profiles indicate the species coverage for the selected species radiation point and contrast the number of species with single-copy members and with multi-copy members.
Related groups
Homologous relations among genes from different orthologous groups identify sets of related orthologous groups delineated for the specific level of the phylogeny. These relations are defined from pairwise Smith–Waterman comparisons between all members of an orthologous group to all members of any related groups with a cutoff of 1e-3. Related groups are identified at each level of the phylogeny-defined hierarchy, linking to ‘sibling orthologous groups’ as opposed to parent or child groups that would correspond to moving up or down the phylogeny.
OrthoDB access
The hierarchy
The phylogeny-defined hierarchy of orthologous groups in OrthoDB allows searches to be performed at specific radiation points by selecting a node of the interactive species trees. Selecting a node encompassing only a few closely related species will focus the search results on more fine-grained orthologous groups of mostly one-to-one relations. Moving toward the root of the tree will include more distantly related species and will generally retrieve more inclusive orthologous groups that contain all the descendants of the ancestral gene.
Text searches
OrthoDB enables relevant data retrieval through specific queries using protein, gene, InterPro or GO identifiers, or more general searches with keywords, names or synonyms, descriptor terms or phrases. Gene annotations sourced from UniProt, supplemented with data from specific resources for representative model organisms from each lineage, provide rich annotations that facilitate comprehensive database searching. The text search feature provides additional flexibility using simple logical operator syntax to build complex queries; e.g. to optionally include variations of a term, or to exclude terms (Supplementary Table S3). In addition, specific protein domain architectures may be queried with a comma-separated N- to C-terminus ordered list of InterPro identifiers.
Copy-number profiles
OrthoDB features the ability to search for orthologous groups with specific phyletic profiles to retrieve groups matching specific copy-number criteria such as all single-copy or all multi-copy orthologs. Combining the criteria of absent, present, single-copy, multi-copy or no restriction, for each species within a selected clade can generate numerous variations of user-defined phyletic profiles for database querying. These profile query options are extended through a selection of predefined common profiles with more relaxed search criteria, e.g. single-copy orthologs but allowing for a gene loss or duplication event in one species.
Sequence similarity
The BLAST search facility ensures that data interrogation is not limited by the coverage of detailed gene annotations. The relevant data of the orthologous group closest to the root-level are returned if a protein sequence match with a significant BLAST hit is identified. Such sequence-based queries help to circumvent potential ambiguities arising from multiple gene identifiers or synonyms from alternative resources or database releases.
Query history
Queries are stored during each user’s web browser session to enable reviewing and re-running of their recently executed queries. The type of search and the level in the species phylogeny at which it was performed, together with the number of orthologous groups returned, are displayed for each query. The user may re-run or delete individual queries or clear their complete query history.
Data export
The data for each orthologous group may be exported as either a Fasta-formatted file of protein sequences or a tab-delimited text file of members with their InterPro annotations. In addition, data for the complete set of groups retrieved from any OrthoDB query may be exported as both Fasta-formatted sequence and tab-delimited annotation files. The ‘Print Tables’ option exports the data tables for all retrieved groups to a printer-friendly HTML-formatted document that may be printed or saved as required.
OrthoDB links
OrthoDB data are cross-referenced with numerous biological databases, linking retrieved orthologous group gene members to their respective sources and allowing direct access to additional information. In addition, OrthoDB groups are referenced through link-outs from major community resources including UniProt and Flybase.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The Swiss National Science Foundation (31003A-125350). Funding for open access charge: Swiss Institute of Bioinformatics.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Dr Ivo Pedruzzi and all members of the Computational Evolutionary Genomics Group for useful suggestions and discussions.
REFERENCES
- 1.Fitch W. Distinguishing homologous from analogous proteins. Syst. Zool. 1970;19:99–113. [PubMed] [Google Scholar]
- 2.Koonin E. Orthologs, paralogs, and evolutionary genomics. Annu. Rev. Genet. 2005;39:309–338. doi: 10.1146/annurev.genet.39.073003.114725. [DOI] [PubMed] [Google Scholar]
- 3.Sonnhammer E, Koonin E. Orthology, paralogy and proposed classification for paralog subtypes. Trends Genet. 2002;18:619–620. doi: 10.1016/s0168-9525(02)02793-2. [DOI] [PubMed] [Google Scholar]
- 4.Fang G, Bhardwaj N, Robilotto R, Gerstein M. Getting started in gene orthology and functional analysis. PLoS Comput. Biol. 2010;6:e1000703. doi: 10.1371/journal.pcbi.1000703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Altenhoff A, Dessimoz C. Phylogenetic and functional assessment of orthologs inference projects and methods. PLoS Comput. Biol. 2009;5:e1000262. doi: 10.1371/journal.pcbi.1000262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Tatusov R, Fedorova N, Jackson J, Jacobs A, Kiryutin B, Koonin E, Krylov D, Mazumder R, Mekhedov S, Nikolskaya A, et al. The COG database: an updated version includes eukaryotes. BMC Bioinformatics. 2003;4:41. doi: 10.1186/1471-2105-4-41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Muller J, Szklarczyk D, Julien P, Letunic I, Roth A, Kuhn M, Powell S, von Mering C, Doerks T, Jensen L, et al. eggNOG v2.0: extending the evolutionary genealogy of genes with enhanced non-supervised orthologous groups, species and functional annotations. Nucleic Acids Res. 2010;38:D190–D195. doi: 10.1093/nar/gkp951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ostlund G, Schmitt T, Forslund K, Köstler T, Messina D, Roopra S, Frings O, Sonnhammer E. InParanoid 7: new algorithms and tools for eukaryotic orthology analysis. Nucleic Acids Res. 2010;38:D196–D203. doi: 10.1093/nar/gkp931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Roth A, Gonnet G, Dessimoz C. Algorithm of OMA for large-scale orthology inference. BMC Bioinformatics. 2008;9:518. doi: 10.1186/1471-2105-9-518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kriventseva E, Rahman N, Espinosa O, Zdobnov E. OrthoDB: the hierarchical catalog of eukaryotic orthologs. Nucleic Acids Res. 2008;36:D271–D275. doi: 10.1093/nar/gkm845. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, Mackey A, Stoeckert CJ, Roos D. OrthoMCL-DB: querying a comprehensive multi-species collection of ortholog groups. Nucleic Acids Res. 2006;34:D363–D368. doi: 10.1093/nar/gkj123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Deluca T, Wu I, Pu J, Monaghan T, Peshkin L, Singh S, Wall D. Roundup: a multi-genome repository of orthologs and evolutionary distances. Bioinformatics. 2006;22:2044–2046. doi: 10.1093/bioinformatics/btl286. [DOI] [PubMed] [Google Scholar]
- 13.Vilella A, Severin J, Ureta-Vidal A, Heng L, Durbin R, Birney E. EnsemblCompara GeneTrees: complete, duplication-aware phylogenetic trees in vertebrates. Genome Res. 2009;19:327–335. doi: 10.1101/gr.073585.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van der Heijden R, Snel B, van Noort V, Huynen M. Orthology prediction at scalable resolution by phylogenetic tree analysis. BMC Bioinformatics. 2007;8:83. doi: 10.1186/1471-2105-8-83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas P. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res. 2010;38:D204–D210. doi: 10.1093/nar/gkp1019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Datta R, Meacham C, Samad B, Neyer C, Sjölander K. Berkeley PHOG: PhyloFacts orthology group prediction web server. Nucleic Acids Res. 2009;37:W84–W89. doi: 10.1093/nar/gkp373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huerta-Cepas J, Bueno A, Dopazo J, Gabaldón T. PhylomeDB: a database for genome-wide collections of gene phylogenies. Nucleic Acids Res. 2008;36:D491–D496. doi: 10.1093/nar/gkm899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wapinski I, Pfeffer A, Friedman N, Regev A. Automatic genome-wide reconstruction of phylogenetic gene trees. Bioinformatics. 2007;23:i549–i558. doi: 10.1093/bioinformatics/btm193. [DOI] [PubMed] [Google Scholar]
- 19.Ruan J, Li H, Chen Z, Coghlan A, Coin L, Guo Y, Hériché J, Hu Y, Kristiansen K, Li R, et al. TreeFam: 2008 update. Nucleic Acids Res. 2008;36:D735–D740. doi: 10.1093/nar/gkm1005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Duret L, Mouchiroud D, Gouy M. HOVERGEN: a database of homologous vertebrate genes. Nucleic Acids Res. 1994;22:2360–2365. doi: 10.1093/nar/22.12.2360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Tatusov R, Koonin E, Lipman D. A genomic perspective on protein families. Science. 1997;278:631–637. doi: 10.1126/science.278.5338.631. [DOI] [PubMed] [Google Scholar]
- 22.Hahn M. Distinguishing among evolutionary models for the maintenance of gene duplicates. J. Hered. 2009;100:605–617. doi: 10.1093/jhered/esp047. [DOI] [PubMed] [Google Scholar]
- 23.Wilson C, Kreychman J, Gerstein M. Assessing annotation transfer for genomics: quantifying the relations between protein sequence, structure and function through traditional and probabilistic scores. J. Mol. Biol. 2000;297:233–249. doi: 10.1006/jmbi.2000.3550. [DOI] [PubMed] [Google Scholar]
- 24.UniProt-Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tweedie S, Ashburner M, Falls K, Leyland P, McQuilton P, Marygold S, Millburn G, Osumi-Sutherland D, Schroeder A, Seal R, et al. FlyBase: enhancing Drosophila Gene Ontology annotations. Nucleic Acids Res. 2009;37:D555–D559. doi: 10.1093/nar/gkn788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Smith T, Waterman M. Identification of common molecular subsequences. J. Mol. Biol. 1981;147:195–197. doi: 10.1016/0022-2836(81)90087-5. [DOI] [PubMed] [Google Scholar]
- 27.Saebø P, Andersen S, Myrseth J, Laerdahl J, Rognes T. PARALIGN: rapid and sensitive sequence similarity searches powered by parallel computing technology. Nucleic Acids Res. 2005;33:W535–W539. doi: 10.1093/nar/gki423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Richards S, Gibbs R, Weinstock G, Brown S, Denell R, Beeman R, Gibbs R, Bucher G, Friedrich M, Grimmelikhuijzen C, et al. The genome of the model beetle and pest Tribolium castaneum. Nature. 2008;452:949–955. doi: 10.1038/nature06784. [DOI] [PubMed] [Google Scholar]
- 29.Elsik C, Tellam R, Worley K, Gibbs R, Muzny D, Weinstock G, Adelson D, Eichler E, Elnitski L, Guigó R, et al. The genome sequence of taurine cattle: a window to ruminant biology and evolution. Science. 2009;324:522–528. doi: 10.1126/science.1169588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kirkness E, Haas B, Sun W, Braig H, Perotti M, Clark J, Lee S, Robertson H, Kennedy R, Elhaik E, et al. Genome sequences of the human body louse and its primary endosymbiont provide insights into the permanent parasitic lifestyle. Proc. Natl Acad. Sci. USA. 2010;107:12168–12173. doi: 10.1073/pnas.1003379107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Werren J, Richards S, Desjardins C, Niehuis O, Gadau J, Colbourne J, Beukeboom L, Desplan C, Elsik C, Grimmelikhuijzen C, et al. Functional and evolutionary insights from the genomes of three parasitoid Nasonia species. Science. 2010;327:343–348. doi: 10.1126/science.1178028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Waterhouse R, Kriventseva E, Meister S, Xi Z, Alvarez K, Bartholomay L, Barillas-Mury C, Bian G, Blandin S, Christensen B, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316:1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Wyder S, Kriventseva E, Schröder R, Kadowaki T, Zdobnov E. Quantification of ortholog losses in insects and vertebrates. Genome Biol. 2007;8:R242. doi: 10.1186/gb-2007-8-11-r242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lemay D, Lynn D, Martin W, Neville M, Casey T, Rincon G, Kriventseva E, Barris W, Hinrichs A, Molenaar A, et al. The bovine lactation genome: insights into the evolution of mammalian milk. Genome Biol. 2009;10:R43. doi: 10.1186/gb-2009-10-4-r43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Matsui T, Yamamoto T, Wyder S, Zdobnov E, Kadowaki T. Expression profiles of urbilaterian genes uniquely shared between honey bee and vertebrates. BMC Genomics. 2009;10:17. doi: 10.1186/1471-2164-10-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Flicek P, Aken B, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Coates G, Fairley S, et al. Ensembl's 10th year. Nucleic Acids Res. 2010;38:D557–D562. doi: 10.1093/nar/gkp972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Legeai F, Shigenobu S, Gauthier J, Colbourne J, Rispe C, Collin O, Richards S, Wilson A, Murphy T, Tagu D. AphidBase: a centralized bioinformatic resource for annotation of the pea aphid genome. Insect Mol. Biol. 2010;19(Suppl. 2):5–12. doi: 10.1111/j.1365-2583.2009.00930.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kim H, Murphy T, Xia J, Caragea D, Park Y, Beeman R, Lorenzen M, Butcher S, Manak J, Brown S. BeetleBase in 2010: revisions to provide comprehensive genomic information for Tribolium castaneum. Nucleic Acids Res. 2010;38:D437–D442. doi: 10.1093/nar/gkp807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Duan J, Li R, Cheng D, Fan W, Zha X, Cheng T, Wu Y, Wang J, Mita K, Xiang Z, et al. SilkDB v2.0: a platform for silkworm (Bombyx mori) genome biology. Nucleic Acids Res. 2010;38:D453–D456. doi: 10.1093/nar/gkp801. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lawson D, Arensburger P, Atkinson P, Besansky N, Bruggner R, Butler R, Campbell K, Christophides G, Christley S, Dialynas E, et al. VectorBase: a data resource for invertebrate vector genomics. Nucleic Acids Res. 2009;37:D583–D587. doi: 10.1093/nar/gkn857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Colbourne J, Singan V, Gilbert D. wFleaBase: the Daphnia genome database. BMC Bioinformatics. 2005;6:45. doi: 10.1186/1471-2105-6-45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Bult C, Kadin J, Richardson J, Blake J, Eppig J. The Mouse Genome Database: enhancements and updates. Nucleic Acids Res. 2010;38:D586–D592. doi: 10.1093/nar/gkp880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Engel S, Balakrishnan R, Binkley G, Christie K, Costanzo M, Dwight S, Fisk D, Hirschman J, Hitz B, Hong E, et al. Saccharomyces Genome Database provides mutant phenotype data. Nucleic Acids Res. 2010;38:D433–D436. doi: 10.1093/nar/gkp917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.GO-Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Hunter S, Apweiler R, Attwood T, Bairoch A, Bateman A, Binns D, Bork P, Das U, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Hedges SB, Kumar S. The Timetree of Life. Oxford: Oxford University Press; 2009. [Google Scholar]
- 47.Dujon B. Yeasts illustrate the molecular mechanisms of eukaryotic genome evolution. Trends Genet. 2006;22:375–387. doi: 10.1016/j.tig.2006.05.007. [DOI] [PubMed] [Google Scholar]