Abstract
Phospho3D is a database of three-dimensional (3D) structures of phosphorylation sites (P-sites) derived from the Phospho.ELM database, which also collects information on the residues surrounding the P-site in space (3D zones). The database also provides the results of a large-scale structural comparison of the 3D zones versus a representative dataset of structures, thus associating to each P-site a number of structurally similar sites. The new version of Phospho3D presents an 11-fold increase in the number of 3D sites and incorporates several additional features, including new structural descriptors, the possibility of selecting non-redundant sets of 3D structures and the availability for download of non-redundant sets of structurally annotated P-sites. Moreover, it features P3Dscan, a new functionality that allows the user to submit a protein structure and scan it against the 3D zones collected in the Phospho3D database. Phospho3D version 2.0 is available at: http://www.phospho3d.org/.
INTRODUCTION
During recent years, there has been an increasing interest in the structural features of protein phosphorylation sites (P-sites). This fact can be ascribed to the steadily growing experimentally verified P-sites provided by high-throughput mass spectrometry-based proteomics techniques [e.g. (1)]. The simultaneous availability of an increasing number of three-dimensional (3D) structures is making it possible to infer the structural context for a significant number of P-sites. In order to identify the structural determinants of kinase specificity, some authors tried to characterize the 3D environment of P-sites with the aim of pinpointing specific structural features (2–4). Both phosphorylation sites databases, also reporting structural data, and interesting systematic structural analyses of P-sites have recently appeared in the literature [for a review, see ref. (5)]. PHOSIDA (1), a phosphorylation sites database, includes the predicted accessibility and secondary structure of each P-site. The mtcPTM (6) database stores homology models for proteins and protein domains that contain phosphorylated residues. Finally, many of the P-site predictors incorporating 3D-context information (1,4,7–10) display an improvement in performance with respect to predictors using sequence information only.
So far the structural attributes stored in P-site databases and incorporated in P-site predictors are essentially of two types: accessibility and secondary structure.
In 2007, we presented Phospho3D, a database of reliably predicted 3D structures of protein phosphorylation sites (11), derived from the Phospho.ELM database (12) and enriched with structural annotation at the residue level, including accessibility, secondary structure and residue conservation as from the Consurf-HSSP database (13). Phospho3D also stored and annotated the sequence flanking the P-site (10 residues) and the zone, i.e. the 3D region defined by the set of residues at a distance not exceeding 12 Å from the phospho-instance.
Since then, the number of Phospho.ELM instances increased ∼4-fold, raising from 5314 (in 1805 proteins) to 42 474 (in 8718 proteins), and more than 26 000 structures were added to the Protein Data Bank (PDB) (14).
Here we introduce Phospho3D version 2.0, which—besides an eleven-fold increase in the number of Phospho.ELM unique instances mapped onto 3D structures (compared to version 1.0)—incorporates several new features, including additional structural descriptors of P-sites, the possibility of browsing the database selecting non-redundant sets of 3D structures, the availability for download of many non-redundant sets of structurally annotated P-sites—aimed at serving as reliable benchmark datasets for predictors’ training and test—and P3Dscan, a new functionality that allows the user to submit a protein structure and scan it against the 3D P-site zones collected in the Phospho3D database.
DATABASE CONTENTS
The updated Phospho3D database was constructed by collecting data from the latest release of the Phospho.ELM database (Version 9.0, August 2010), which currently stores about 42 500 experimentally verified phosphorylation sites in 8718 substrate proteins, both manually extracted from the literature and obtained from mass spectrometry-based proteomics experiments. The correspondence between Phospho.ELM sequences and PDB chains was based on sequence alignment using at least 98% sequence identity. P-sites in gapped regions of the alignment were discarded.
This resulted in 5387 mapped instances (1770 unique Phospho.ELM instances on 2158 protein chains—897 Ser, 338 Thr, 535 Tyr).
Notice that P-sites derived from mass spectrometry (MS) experiments should be taken with caution. In fact, due to the current procedures for MS data deposition, it is difficult to systematically detect if a phospho-instance was identified in physiologically abnormal conditions (e.g. in proteins extracted from oncogenic tissues or that do not undergo phosphorylation, such as hemoglobin) (15). In order to help users detect such potentially problematic cases, we reported—for each P-site—the nature of the original experiment (low- or high-throughput) and the corresponding literature reference (PMID). Moreover, we encourage users to carefully analyze the structural context of P-sites, which might be indicative of problems in the original data. One example is represented by the Tyr phosphorylation site mapped to position 133 of the human hemoglobin subunit beta (UniProtKB:P68871), for which Phospho3D stores 43 PDB structures. In most of the reported structures, the solvent accessibility of Y133 is zero and it is never >3.5%. This structural information suggests that the original data might not be reliable.
The basic information stored in Phospho3D consists of the P-site instance, its flanking sequence (10 residues) and the P-site 3D zone, i.e. the set of residues in a 12 Å radius surrounding the P-site in space. For each residue in the zone Phospho3D 2.0 stores the following structural descriptors: secondary structure and solvent accessibility (in Å2) as defined by DSSP (16); percentage solvent accessibility, obtained by normalizing the DSSP solvent accessibility by the maximum accessibility value for each residue as determined in ref. (17); B-factor, computed as specified in ref. (18); occurrence in a cavity together with the rank and volume of the cavity calculated with the SURFNET program (19); the depth index DPX (20) and the protrusion index CX (21), obtained using the PSAIA software (22); the CONSURF conservation score extracted from ConSurfDB (23); the disorder probability provided by DisEMBL (24) according to three different criteria: (i) loops/coils as defined by DSSP, (ii) hot loops, i.e. loops with a high degree of mobility as determined by temperature (B-) factor and (iii) missing coordinates in X-Ray structure as defined by REMARK-465 entries in PDB.
A detailed description of each structural attribute is reported in the website documentation.
Phospho3D 2.0 also provides information derived from Phospho.ELM, such as, when available, the kinase(s) phosphorylating a given P-site, and, for each zone, the results of a large-scale local structural comparison versus a non redundant (sequence identity ≤20%) dataset of 487 PDB X-ray protein chains with experimental resolution ≤1.5 Å extracted from eukaryotic organisms. The comparison is carried out using the new version of the algorithm (25) and the same criteria for assessing structural similarity used in the previous database version (11) although more stringent thresholds are applied in this case, as described in the website documentation. The database queries can now be performed on seven PDB non-redundant sets: the whole collection of P-sites, the set of P-sites found in non-identical structures (PDB100) and P-sites found in PDB structures belonging to five redundancy sets, ranging from PDB90 to PDB20, where the number corresponds to the maximum sequence identity shared by the protein chains in the redundancy set. These sets have been determined using the PISCES resource (26).
Additionally, the P-site annotations at the residue level are available for download on the Phospho3D website. These can serve as benchmark for P-site predictors’ training and test and for analyses of P-site structural features.
Finally, Phospho3D 2.0 now links each entry to the corresponding Phospho.ELM instance and the kinase names to their UniProt ACs (27).
P3DSCAN
Phospho3D 2.0 provides a novel functionality that allows the user to upload a PDB-formatted structure and perform a local structural comparison against the 5387 zones (one for each Phospho.ELM mapped instance) stored in the database, aimed at identifying local structural similarities between the user query structure and one of the structural patches containing a P-site. In order to evaluate the structural context of each match, we provide its graphical display and a table reporting the structural information at the residue level of both the query and the target 3D matching patches. The comparison algorithm—that P3Dscan runs on-the-fly—is the same as the one used for the large-scale comparison whose results are stored in the database. The comparison results are also provided in text format for download.
THE WEB INTERFACE
Similarly to the previous version, Phospho3D 2.0 can be searched by kinase name, by PDB identification code or by keyword. In this new version, however, the user can additionally select a redundancy set in order to avoid retrieving identical or very similar P-sites. The data returned to the user consist of a brief description of the PDB structure(s) that fulfill the search criteria and a list of instances presented along with associated information. In particular, each instance is now linked to the corresponding Phospho.ELM entry. For each P-site, the user can select three options related to the surrounding structural zone: a graphical view using the Jmol Java Applet (http://www.jmol.org), a tabular view reporting the zone annotation at the residue level or a list of 3D matches identified by local structural comparison. Each match can be visualized using Jmol.
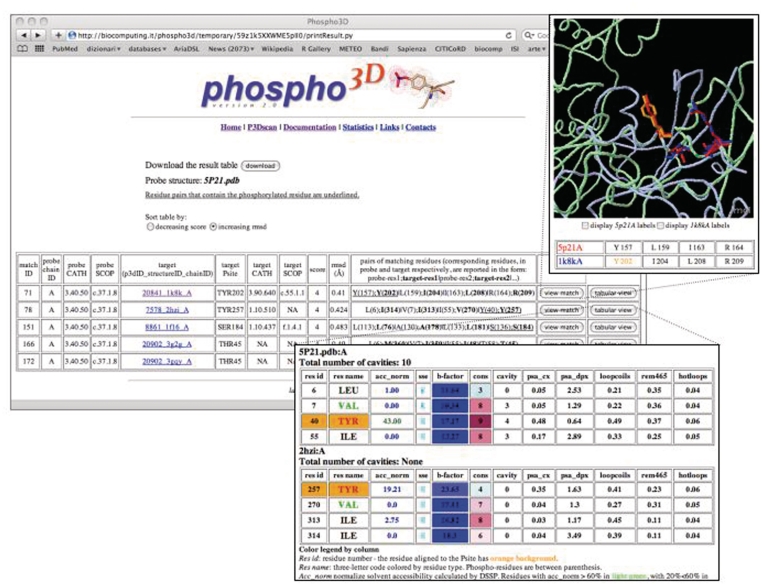
The P3Dscan webpage can be reached from the Phospho3D homepage. Users upload a PDB-formatted file, choose a redundancy set of 3D zones they want to scan against their structure and run the comparison by clicking the ‘p3d scan’ button. P3Dscan results are displayed in tabular format (Figure 1). The result table can be sorted by increasing match score or decreasing RMSD. Each line of the table reports the information of a single match. A match can be graphically visualized by clicking on the corresponding button. Moreover, the tabular view button links to a window displaying structural annotation at the residue level, both for the query and the target 3D patches. The Result Table can also be downloaded in text format.
Figure 1.
The P3Dscan output page for the crystal structure of the H-ras oncogene protein p21 (PDB 5P21). The table reports the list of matches between the query protein (probe) and the zone(s) collected in the database (target). SCOP and CATH annotation, when available, is reported for both the query structure and for each matching zone therefore the user can discriminate matches due to overall fold similarity from functionally interesting ones. The 3D zone (column 5) is linked to the corresponding Phospho3D database entry and the phospho-residue is explicitly reported in column 6. The score (column 9) corresponds to the number of paired residues in the match. The RMSD (column 10) is calculated on the matching amino acids. The pairs of residues participating in the match are reported in column 11. Upper right inset: graphical display (Jmol) of the probe and target structures. Residues participating in the match are in stick and the P-site is colored in orange. Bottom inset: tabular view: structural information at the residue level is reported for both the probe (user query structure) and target (3D zone) residues involved in the match.
STRUCTURAL ANALYSIS
We performed a large-scale structural analysis of the P-sites stored in Phospho3D and plotted the statistical distributions of each 3D attribute used to annotate the P-sites in the database. The analysis was carried out separately for each redundancy set. The distributions for the P-sites falling on non-identical structures (PISCES PDB100) can be found at http://www.phospho3d.org/stats.py#3.
CONCLUSIONS
The new version of Phospho3D stores a markedly increased number of structurally annotated P-sites. In addition, it incorporates new significant improvements, such as several new structural descriptors, non-redundant datasets and a tool, P3Dscan, for the analysis of uploaded protein structures.
We believe that this enhanced version of the database makes it possible to fully exploit available structural information on P-sites and use it to perform structural analyses and/or build P-site predictors.
Importantly, the Phospho3D update procedure is now completely automated, allowing regular and timely updates of the database with each new Phospho.ELM release.
FUNDING
This work was supported by Istituto Pasteur—Fondazione Cenci Bolognetti, Roma; the 7th EC Framework Programme LEISHDRUG project [grant number 223414]; and a ‘Juan de la Cierva’ fellowship to A.Z. Funding for open access charge: 7th EC Framework Programme LEISHDRUG project.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Many thanks to Holger Dinkel (Phospho.ELM database) for technical support.
REFERENCES
- 1.Gnad F, Ren S, Cox J, Olsen JV, Macek B, Oroshi M, Mann M. PHOSIDA (phosphorylation site database): management, structural and evolutionary investigation, and prediction of phosphosites. Genome Biol. 2007;8:R250. doi: 10.1186/gb-2007-8-11-r250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Fan SC, Zhang XG. Characterizing the microenvironment surrounding phosphorylated protein sites. Genomics Proteomics Bioinformatics. 2005;3:213–217. doi: 10.1016/S1672-0229(05)03029-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kitchen J, Saunders RE, Warwicker J. Charge environments around phosphorylation sites in proteins. BMC Struct. Biol. 2008;8:19. doi: 10.1186/1472-6807-8-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Durek P, Schudoma C, Weckwerth W, Selbig J, Walther D. Detection and characterization of 3D-signature phosphorylation site motifs and their contribution towards improved phosphorylation site prediction in proteins. BMC Bioinformatics. 2009;10:117. doi: 10.1186/1471-2105-10-117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Via A, Diella F, Gibson TJ, Helmer-Citterich M. From sequence to structural analysis in protein phosphorylation motifs. Front. Biosci. 2011;16:1261–1275. doi: 10.2741/3787. [DOI] [PubMed] [Google Scholar]
- 6.Jimenez JL, Hegemann B, Hutchins JR, Peters JM, Durbin R. A systematic comparative and structural analysis of protein phosphorylation sites based on the mtcPTM database. Genome Biol. 2007;8:R90. doi: 10.1186/gb-2007-8-5-r90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Blom N, Gammeltoft S, Brunak S. Sequence and structure-based prediction of eukaryotic protein phosphorylation sites. J. Mol. Biol. 1999;294:1351–1362. doi: 10.1006/jmbi.1999.3310. [DOI] [PubMed] [Google Scholar]
- 8.Brinkworth RI, Breinl RA, Kobe B. Structural basis and prediction of substrate specificity in protein serine/threonine kinases. Proc. Natl Acad. Sci. USA. 2003;100:74–79. doi: 10.1073/pnas.0134224100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Plewczynski D, Tkacz A, Godzik A, Rychlewski L. A support vector machine approach to the identification of phosphorylation sites. Cell Mol. Biol. Lett. 2005;10:73–89. [PubMed] [Google Scholar]
- 10.Plewczynski D, Jaroszewski L, Godzik A, Kloczkowski A, Rychlewski L. Molecular modeling of phosphorylation sites in proteins using a database of local structure segments. J. Mol. Model. 2005;11:431–438. doi: 10.1007/s00894-005-0235-z. [DOI] [PubMed] [Google Scholar]
- 11.Zanzoni A, Ausiello G, Via A, Gherardini PF, Helmer-Citterich M. Phospho3D: a database of three-dimensional structures of protein phosphorylation sites. Nucleic Acids Res. 2007;35:D229–D231. doi: 10.1093/nar/gkl922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Diella F, Gould CM, Chica C, Via A, Gibson TJ. Phospho.ELM: a database of phosphorylation sites – update 2008. Nucleic Acids Res. 2008;36:D240–D244. doi: 10.1093/nar/gkm772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Glaser F, Rosenberg Y, Kessel A, Pupko T, Ben-Tal N. The ConSurf-HSSP database: the mapping of evolutionary conservation among homologs onto PDB structures. Proteins. 2005;58:610–617. doi: 10.1002/prot.20305. [DOI] [PubMed] [Google Scholar]
- 14.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nichols AM, White FM. Manual validation of peptide sequence and sites of tyrosine phosphorylation from MS/MS spectra. Methods Mol. Biol. 2009;492:143–160. doi: 10.1007/978-1-59745-493-3_8. [DOI] [PubMed] [Google Scholar]
- 16.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 17.Miller S, Janin J, Lesk AM, Chothia C. Interior and surface of monomeric proteins. J. Mol. Biol. 1987;196:641–656. doi: 10.1016/0022-2836(87)90038-6. [DOI] [PubMed] [Google Scholar]
- 18.Bartlett GJ, Porter CT, Borkakoti N, Thornton JM. Analysis of catalytic residues in enzyme active sites. J. Mol. Biol. 2002;324:105–121. doi: 10.1016/s0022-2836(02)01036-7. [DOI] [PubMed] [Google Scholar]
- 19.Laskowski RA. SURFNET: a program for visualizing molecular surfaces, cavities, and intermolecular interactions. J. Mol. Graph. 1995;13:323–330, 307–328. doi: 10.1016/0263-7855(95)00073-9. [DOI] [PubMed] [Google Scholar]
- 20.Pintar A, Carugo O, Pongor S. DPX: for the analysis of the protein core. Bioinformatics. 2003;19:313–314. doi: 10.1093/bioinformatics/19.2.313. [DOI] [PubMed] [Google Scholar]
- 21.Pintar A, Carugo O, Pongor S. CX, an algorithm that identifies protruding atoms in proteins. Bioinformatics. 2002;18:980–984. doi: 10.1093/bioinformatics/18.7.980. [DOI] [PubMed] [Google Scholar]
- 22.Mihel J, Sikic M, Tomic S, Jeren B, Vlahovicek K. PSAIA - protein structure and interaction analyzer. BMC Struct. Biol. 2008;8:21. doi: 10.1186/1472-6807-8-21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goldenberg O, Erez E, Nimrod G, Ben-Tal N. The ConSurf-DB: pre-calculated evolutionary conservation profiles of protein structures. Nucleic Acids Res. 2009;37:D323–D327. doi: 10.1093/nar/gkn822. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Linding R, Jensen LJ, Diella F, Bork P, Gibson TJ, Russell RB. Protein disorder prediction: implications for structural proteomics. Structure. 2003;11:1453–1459. doi: 10.1016/j.str.2003.10.002. [DOI] [PubMed] [Google Scholar]
- 25.Gherardini PF, Ausiello G, Helmer-Citterich M. Superpose3D: a local structural comparison program that allows for user-defined structure representations. PLoS One. 2010;5:e11988. doi: 10.1371/journal.pone.0011988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wang G, Dunbrack RL., Jr PISCES: a protein sequence culling server. Bioinformatics. 2003;19:1589–1591. doi: 10.1093/bioinformatics/btg224. [DOI] [PubMed] [Google Scholar]
- 27.The Universal Protein Resource (UniProt) (2010) Nucleic Acids Res. 38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]