Abstract
The Pancreatic Expression database (PED, http://www.pancreasexpression.org) has established itself as the main repository for pancreatic-derived -omics data. For the past 3 years, its data content and access have increased substantially. Here we describe several of its new and improved features, such as data content, which now includes over 60 000 measurements derived from transcriptomics, proteomics, genomics and miRNA profiles from various pancreas-centred reports on a broad range of specimen and experimental types. We also illustrate the capabilities of its interface, which allows integrative queries that can combine PED data with a growing number of biological resources such as NCBI, Ensembl, UniProt and Reactome. Thus, PED is capable of retrieving and integrating different types of -omics, annotations and clinical data. We also focus on the importance of data sharing and interoperability in the cancer field, and the integration of PED into the International Cancer Genome Consortium (ICGC) data portal.
INTRODUCTION
Pancreatic cancer is the fourth leading cause of cancer-related death world-wide (1) with surgical intervention and radiotherapy having a minimal impact on 5-year survival rates. As a result, patient survival rates have remained relatively unchanged over the past 30 years. Because of the poor prognosis associated with pancreatic cancer, a multitude of studies have been dedicated to elucidating the pathogenesis of this malignancy (2). Augmented by advances in high-throughput technologies, this has resulted in a plethora of -omics data. Despite the magnitude of information available, the heterogeneity and isolation of public datasets prevents researchers from effectively mining, extracting and integrating relevant data into their current research.
The publicly available Pancreatic Expression database (PED) was developed to overcome these obstacles by enabling complex pancreatic datasets to be manipulated, mined and integrated with ease (3). Since its inception in 2007, PED has undergone extensive improvements. The range of clinical and -omics data types available for queries has broadened substantially, facilitating the systematic study of pancreatic cancer.
Unlike many cancer databases specialised in providing single-type information, PED stores four different kinds of -omics data: transcriptomics, proteomics, miRNA and genomics. These profiles are derived from a broad range of specimens from tissues and body fluids of healthy people or patients, cell lines and mouse models as well as different treatments and drugs. This is key to providing a comprehensive overview of the molecular changes in cancer. Another important feature of PED is that it allows for its data to be interrogated from major gene repositories such as NCBI EntrezGene (4) and Ensembl GeneView (5), third-party software such as R statistical environment (6) and Cytoscape (7) or jointly with data from major biological resources such as the Reactome Pathway project (8), PRIDE (9) and UniProt (10). Increased functionality also allows for greater interoperability with international cancer efforts, such as the International Cancer Genome Consortium (ICGC, http://www.icgc.org), a major international collaboration designed to identify the key genetic mutations involved in up to 50 types of cancer, which will enable the development of new and better ways of diagnosing, treating and preventing cancer (11).
To the best of our knowledge, there are neither tools nor databases that provide the same information for cancer research as our platform. By allowing for integration and mining of published pancreatic cancer data in the context of a wide range of annotations, PED offers more options than raw data repositories such as Gene Expression Omnibus (GEO) (12) or ArrayExpress (13). One of the main strengths of our system is the possibility of setting many filters to ask very specific pancreatic cancer-related questions and obtain a focused annotated data output. Here we outline details of the improved data content, query interfaces and interoperability.
IMPROVED DATA CONTENT
The database contains 56 015 differential or expression measurements and 6363 DNA copy number alterations. These data values are extracted from over 59 published studies and include profiles from a large number of specimen types, such as pancreatic tissues and various body fluids obtained from healthy subjects and patients with benign, pre-malignant and malignant diseases. In addition, studies using pancreatic cancer cell lines and murine models have also been incorporated (Supplementary Table S1). Where applicable, information on the different treatment conditions applied is provided to users.
The collected samples were profiled on a wide range of transcriptomics, proteomics, miRNAs and genomics platforms (Supplementary Table S2). To date, PED describes pancreatic-related regulation events in 8229 genes/proteins, 27 327 transcripts and 279 miRNA as well as 2771 gains, 1073 losses, 347 homozygous deletions, 1297 high-level amplifications and 875 loss of heterozygosity events occurring in distinct genomic areas.
The data collection pipeline from the original database was enhanced to cover not only transcriptomics but also genomics, proteomics and miRNA information. Data from the primary literature were manually curated, reviewed for accuracy and consistency, and loaded into a relational database. The Ensembl annotations originally used for mapping probe identifiers and gene names to standard values (version 48) have been updated to a more current version (version 56). This ensures that our database remains up-to-date with ongoing improvements to annotation and microarray probe set mappings and helps avoid data integrity errors. The data collection process was expanded to encompass new data types such as DNA copy number changes. Here, chromosomal coordinates for each copy number variation identified from papers were mapped to genome release GRCh37 with conversions between genome versions carried out using the liftover tool from UCSC (14).
We imported the available Ensembl human gene annotations (5) for genes, proteins, SNP information, sequences, gene structure and multi-species data, enabling the integration and annotation of heterogeneous pancreatic data (Supplementary Table S3).
IMPROVED QUERY CAPABILITIES
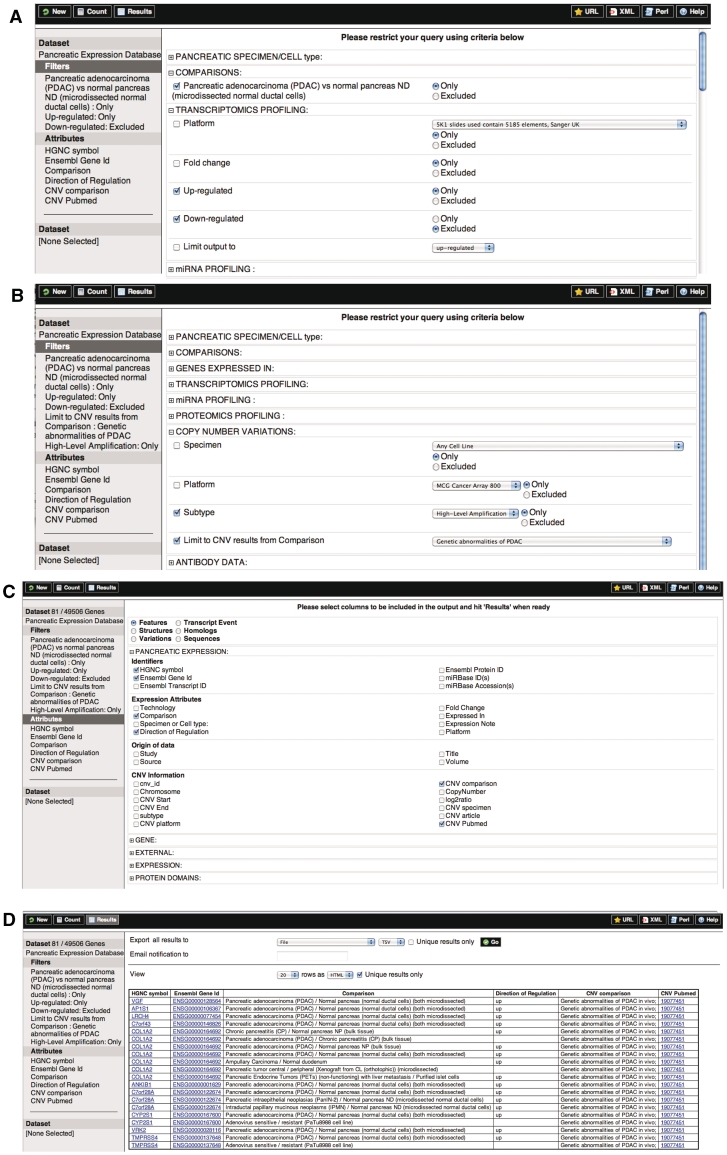
While the original PED system included tools for querying across different tumour stages of pancreatic cancer and different pancreatic disease types in a simple integrated way (3), the interface has been greatly improved to provide the user access to the expanded data content. By including new platforms, comparisons and data types, it is possible to combine information and, therefore, to perform more complex queries than by viewing expression data alone. For example, it is possible to highlight the impact of copy number aberrations on gene expression patterns to filter out genes whose expression levels are not consistent with their DNA copy number status and point to the subset of candidate oncogenes or tumour suppressor genes showing copy number-driven expression changes (Figure 1). There are now options available for proteomics, transcriptomics and miRNA profiling, allowing these data types to be queried in isolation or combined to look for genes consistently identified across different data technologies or to extract specific data such as microRNAs deregulated in the different stages of pancreatic cancer.
Figure 1.
Integrated view of genomic and transcriptomic changes in pancreatic cancer. This figure integrates the results from both genome-wide DNA copy number and genome-wide gene expression profiling. In a few seconds, this allows for visual inspection of copy number-driven expression changes. Here the query is to find genes differentially up-regulated in PDAC versus normal using microdissected ductal cells (A) and then combine this information with data on genes that are also associated with genomic variations in PDAC samples by combining with results for copy number changes on high level amplifications (B). Pick attributes for display (C). A summary of the first 20 results is shown in (D).
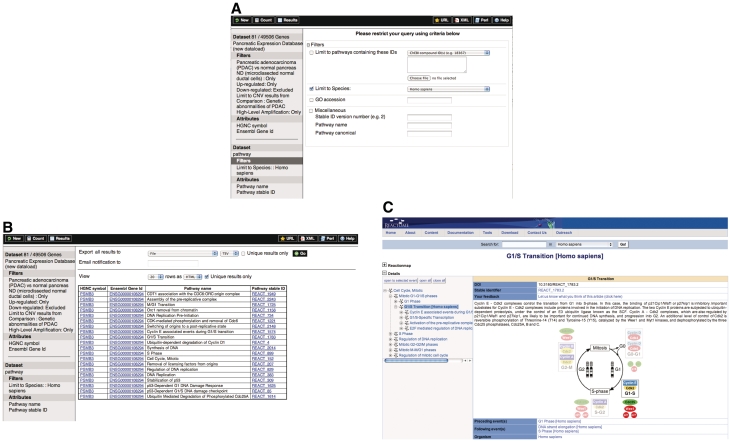
The new interface incorporates the full functionality and data from Ensembl Mart 56 including richer data content and more advanced querying capabilities and also makes better use of the BioMart interface capabilities (15) for cross-linking different datasets that have data types in common; for example by combining PED data with Reactome pathway data of human reactions and pathways (8) (Figure 2). This integration allows researchers to quickly identify both pancreatic cancer genes and the pathways they control.
Figure 2.
Cross-linking pathways information from reactome. Results from the previous query (shown in Figure 1) can be further mined and merged with data from Reactome. Here we select Reactome pathways as a second dataset to combine in queries and then restrict obtained information to ‘Homo sapiens’ data (A). Pick attributes and display a summary of the first 20 results (B). Query results return pathway name and stable ID with a hyper-link to the Reactome website allowing to instantly extract data (C).
IMPROVED DATA ACCESS
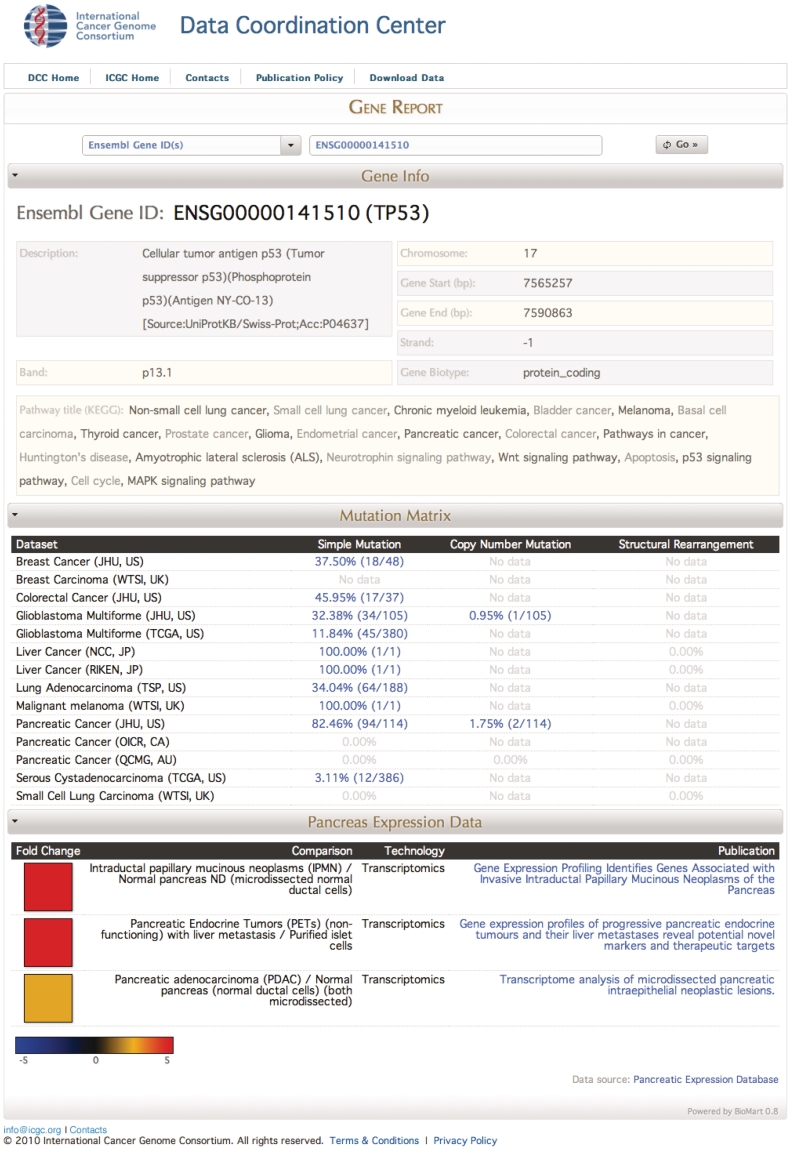
PED is freely accessible through a BioMart web-based query interface at http://www.pancreasexpression.org. PED is also a DAS server (16) providing DAS annotations for the wider community, so it can be used in other resources or browsers such as Ensembl GeneView using the GeneDAS protocol. Access is also possible via web services through third party software tools that have been made compatible with BioMart resources such as Bioconductor (http://www.bioconductor.org) (6,17), Galaxy (18) and Cytoscape (7). Interoperability with the ICGC is also possible through this web services layer and PED is now available through the ICGC data portal (http://dcc.icgc.org) (Figure 3) allowing researchers to conduct combined queries on ICGC experimental data alongside PED literature data. This further increases the ways in which the database can be accessed and its exposure to a variety of disciplines and interests in the scientific community. The database is also accessible as a Linkout resource from NCBI EntrezGene (4). This allows EntrezGene users to be alerted to pancreatic expression data by the presence of a data link for relevant genes that are in the database.
Figure 3.
Interoperability with the ICGC experimental data. Access available from http://dcc.icgc.org. The figure shows the ICGC data portal report for the TP53 gene including gene information; ICGC experimental sequencing results obtained from the participating centres; and PED data. In the PED Report section, a heatmap represents, visually, the level of de-regulation as extracted from the original publication and stored in PED. If you hold your mouse over the coloured box, it will display the fold change value (when available).
DISCUSSION
We have described how PED has evolved from its original role as a repository for cancer transcriptomics data into a comprehensive resource capable of providing a quick overview of molecular changes at the transcriptome, proteome, genome and/or miRNA level. Consequently, there has been a huge growth in its -omics, specimen/clinical and annotation data content.
We believe that interoperability is a key factor in the utility and productive use of any current and future cancer databases. This is essential to ensure the sustainability of any cancer database and facilitate its integration with major international efforts in cancer research such as the ICGC. This will also allow the design and implementation of more sophisticated analysis portals. The cancer research community needs open source, fully interoperable resources allowing information connectivity and data sharing. Only these types of resource can ensure that cancer data generated across different organisations are shared, thereby maximising the impact of cancer research. By using the BioMart technology for its data management system, PED is fully interoperable with the ICGC. This ensures that PED is integrated with The Cancer Genome Atlas (TCGA, http://cgap.nci.nih.gov) data available through the ICGC data portal. The BioMart web service layer also allows PED to be integrated with several other data sources that also use the BioMart technology such as Reactome, PRIDE, UniProt and Ensembl. Moreover, PED is a Linkout resource integrated with NCBI.
Our database fills the urgent requirement of the pancreatic cancer community for resources capable of integrating the overflowing influx of data generated by novel high-throughput technologies.
The architectural flexibility of PED is easily extendable to other disease types, with this model being used to create a similar resource for malignant (breast cancer) and non-malignant (neurodegenerative) diseases. Reuse of a similar database design will facilitate complex query capabilities across multiple diseases and data types.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Cancer Research UK (programme grant C355/A6253) and FW6 EU project MolDiag-Paca. R.C. is funded by Breast Cancer Campaign. Funding for open access charge: Cancer Research UK.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank our colleagues who have suggested or contributed data for current or previous versions of the database. They also would like to thank the swift and helpful advice from the ICGC DCC and BioMart team.
REFERENCES
- 1.Hariharan D, Saied A, Kocher HM. Analysis of mortality rates for pancreatic cancer across the world. HPB. 2008;10:58–62. doi: 10.1080/13651820701883148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Goonetilleke KS, Siriwardena AK. Current status of gene expression profiling of pancreatic cancer. Int. J. Surg. 2008;6:81–83. doi: 10.1016/j.ijsu.2006.09.001. [DOI] [PubMed] [Google Scholar]
- 3.Chelala C, Hahn SA, Whiteman HJ, Barry S, Hariharan D, Radon TP, Lemoine NR, Crnogorac-Jurcevic T. Pancreatic Expression database: a generic model for the organization, integration and mining of complex cancer datasets. BMC Genomics. 2007;8:439. doi: 10.1186/1471-2164-8-439. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tatusova T. Genomic databases and resources at the National Center for Biotechnology Information. Methods Mol. Biol. 609:17–44. doi: 10.1007/978-1-60327-241-4_2. [DOI] [PubMed] [Google Scholar]
- 5.Hubbard TJ, Aken BL, Ayling S, Ballester B, Beal K, Bragin E, Brent S, Chen Y, Clapham P, Clarke L, et al. Ensembl. Nucleic Acids Res. 2009;37:D690–D697. doi: 10.1093/nar/gkn828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.R Development Core Team. R: a language and environment for statistical computing. R Foundation for Statistical Computing, Vienna, Austria. 2009 http://www.R-project.org. [Google Scholar]
- 7.Cline MS, Smoot M, Cerami E, Kuchinsky A, Landys N, Workman C, Christmas R, Avila-Campilo I, Creech M, Gross B, et al. Integration of biological networks and gene expression data using Cytoscape. Nat. Protoc. 2007;2:2366–2382. doi: 10.1038/nprot.2007.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, Jassal B, et al. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res. 2009;37:D619–D622. doi: 10.1093/nar/gkn863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Jones P, Cote RG, Cho SY, Klie S, Martens L, Quinn AF, Thorneycroft D, Hermjakob H. PRIDE: new developments and new datasets. Nucleic Acids Res. 2008;36:D878–D883. doi: 10.1093/nar/gkm1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.International network of cancer genome projects. Nature. 2010;464:993–998. doi: 10.1038/nature08987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Marshall KA, et al. NCBI GEO: archive for high-throughput functional genomic data. Nucleic Acids Res. 2009;37:D885–D890. doi: 10.1093/nar/gkn764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Parkinson H, Kapushesky M, Kolesnikov N, Rustici G, Shojatalab M, Abeygunawardena N, Berube H, Dylag M, Emam I, Farne A, et al. ArrayExpress update–from an archive of functional genomics experiments to the atlas of gene expression. Nucleic Acids Res. 2009;37:D868–D872. doi: 10.1093/nar/gkn889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rhead B, Karolchik D, Kuhn RM, Hinrichs AS, Zweig AS, Fujita PA, Diekhans M, Smith KE, Rosenbloom KR, Raney BJ, et al. The UCSC Genome Browser database: update 2010. Nucleic Acids Res. 38:D613–D619. doi: 10.1093/nar/gkp939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Haider S, Ballester B, Smedley D, Zhang J, Rice P, Kasprzyk A. BioMart Central Portal – unified access to biological data. Nucleic Acids Res. 2009;37:W23–W27. doi: 10.1093/nar/gkp265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Durinck S, Moreau Y, Kasprzyk A, Davis S, De Moor B, Brazma A, Huber W. BioMart and Bioconductor: a powerful link between biological databases and microarray data analysis. Bioinformatics. 2005;21:3439–3440. doi: 10.1093/bioinformatics/bti525. [DOI] [PubMed] [Google Scholar]
- 18.Blankenberg D, Von Kuster G, Coraor N, Ananda G, Lazarus R, Mangan M, Nekrutenko A, Taylor J. Galaxy: a web-based genome analysis tool for experimentalists. Curr. Protoc. Mol. Biol. doi: 10.1002/0471142727.mb1910s89. Chapter 19, Unit 19.10.1–19.10.21. [DOI] [PMC free article] [PubMed] [Google Scholar]