Abstract
As a reversible post-translational modification (PTM) discovered decades ago, protein lysine acetylation was known for its regulation of transcription through the modification of histones. Recent studies discovered that lysine acetylation targets broad substrates and especially plays an essential role in cellular metabolic regulation. Although acetylation is comparable with other major PTMs such as phosphorylation, an integrated resource still remains to be developed. In this work, we presented the compendium of protein lysine acetylation (CPLA) database for lysine acetylated substrates with their sites. From the scientific literature, we manually collected 7151 experimentally identified acetylation sites in 3311 targets. We statistically studied the regulatory roles of lysine acetylation by analyzing the Gene Ontology (GO) and InterPro annotations. Combined with protein–protein interaction information, we systematically discovered a potential human lysine acetylation network (HLAN) among histone acetyltransferases (HATs), substrates and histone deacetylases (HDACs). In particular, there are 1862 triplet relationships of HAT-substrate-HDAC retrieved from the HLAN, at least 13 of which were previously experimentally verified. The online services of CPLA database was implemented in PHP + MySQL + JavaScript, while the local packages were developed in JAVA 1.5 (J2SE 5.0). The CPLA database is freely available for all users at: http://cpla.biocuckoo.org.
INTRODUCTION
There are two types of acetylation processes widely occurred in proteins (1–8). The first Nα-terminal acetylation is catalyzed a variety of N-terminal acetyltransferases (NATs), which co-translationally transfer acetyl moieties from acetyl-coenzyme A (Acetyl-CoA) to the α-amino (Nα) group of protein amino-terminal residues (1,2). Although Nα-terminal acetylation is rare in prokaryotes, it was estimated that ∼85% of eukaryotic proteins are Nα-terminally modified (1,2). The second type is Nε-lysine acetylation, which specifically modifies ε-amino group of protein lysine residues (3–8). Although Nε-lysine acetylation is less common, it’s one of the most important and ubiquitous post-translational modifications (PTMs) conserved in prokaryotes and eukaryotes (1,2). Moreover, the acetylation and deacetylation are dynamically and temporally regulated by histone acetyltransferases (HATs) and histone deacetylases (HDACs), respectively (4–8).
In 1964, Allfrey et al. (9) first observed that lysine acetylation of histones plays an essential role in regulation of gene expression. Later and recent studies in epigenetics solidified this seminal discovery, and proposed acetylation as a key component of the ‘histone code’ (6). Beyond histones, a wide-range of non-histone proteins can also be lysine acetylated, and involved in a variety of biological processes, such as transcription regulation (10), DNA replication (11), cellular signaling (12), stress response (13) and so on. Aberrance of lysine acetylation and deacetylation is associated with various diseases and cancers (5,7,14). In particular, acetylation was demonstrated to be implicated in cellular metabolism and aging (15–17), while one class of NAD+ dependent HDACs of sirtuins might be potent drug target for promoting longevity (13,17).
Although a great number of efforts have been carried out during the past four decades, the functional contents of lysine acetylation are still far from fully understood. In this regard, identification of acetylated substrates with their sites is fundamental for understanding the molecular mechanisms and regulatory roles of acetylation. In contrast with labor-intensive and time-consuming conventional experimental approaches, recent progresses in acetylome with high-throughput mass spectrometry (MS) have detected thousands of acetylation sites. In 2006, Kim et al. (14) performed a large-scale identification of acetylome with an anti-acetyllysine antibody. There were 195 acetylated proteins with 388 sites detected in HeLa cells and mouse liver mitochondria (14). With a similar strategy, Choudhary et al. (11) experimentally identified 3600 acetylation sites in human. In 2010, Zhao et al. (16) discovered 1047 acetylated substrates in human liver, and demonstrated acetylation playing a major role in metabolic regulation. Furthermore, two acetylomic studies revealed that the functions of lysine acetylation are conserved in Escherichia coli (18) and Salmonella enterica (15).
Since the number of known acetylation sites has rapidly increased, it is an urgent topic to collect the experimental data and provide an integrated resource for the community. Recently, several public databases, such as PhosphoSitePlus (19), HPRD (20), SysPTM (21) and dbPTM (22), have already contained protein acetylation information. In these databases, both of Nα-terminal and Nε-lysine acetylation data were curated, while lysine acetylation sites are usually only a limited part of total sites. For example, SysPTM 1.1 contains 3001 acetylation sites in 2000 proteins, with only 345 lysine sites (∼11.5%) in 397 substrates (21). In dbPTM 2.0, 2071 experimentally verified acetylation sites were collected in 1525 proteins, with only 792 lysine sites (∼38.2%) in 299 targets (22). Interestingly, HPRD release 9 contains 4691 total sites in 1987 proteins, with 4420 lysine sites (∼94.2%) in 1821 substrates (20). However, HPRD database only focuses on human protein information (20), while thousands of lysine acetylation sites in other species still remain to be collected.
With the motivation to meet the desire for complete acetylomes, here we developed a novel database of compendium of protein lysine acetylation (CPLA). From the scientific literature in PubMed, we manually curated 3311 acetylated proteins with 7151 lysine sites (Table 1). In CPLA database, the primary references and other annotations of these substrates were provided, while the protein–protein interaction (PPI) information was also integrated. Based on the Gene Ontology (GO) and InterPro annotations, we carried out an analysis of functional diversities and regulatory roles of lysine acetylation. As 75.64% of total lysine acetylation sites are taken from Homo sapiens, a potential human lysine acetylation network (HLAN) among HATs, substrates and HDACs was constructed, with 1019 PPIs among 199 proteins. Interestingly, we revealed 1862 potential triplet relationships of HAT-substrate-HDAC, while at lease 13 were previously experimentally verified. Taken together, the CPLA database might be an integrated resource for protein lysine acetylation and provide useful information for further experimental or computational considerations.
Table 1.
The data statistics of lysine acetylated proteins in CPLA database
| Organism | Sub.a | Siteb | Per.c (%) |
|---|---|---|---|
| Homo sapiens | 2585 | 5409 | 75.64 |
| Salmonella typhimurium | 190 | 260 | 3.64 |
| Mus musculus | 187 | 552 | 7.72 |
| Escherichia coli | 149 | 259 | 3.62 |
| Saccharomyces cerevisiae | 44 | 85 | 1.19 |
| Others | 156 | 586 | 8.20 |
| Total | 3311 | 7151 | 100 |
aSub., number of substrates; bSite, number of acetylation sites; cPer., percentiles of acetylation sites.
CONSTRUCTION AND CONTENT
To ensure the quality of CPLA database, we searched the PubMed with a major keyword ‘acetylation’ and collected experimentally identified lysine acetylated proteins with their sites from more than 18 500 published articles (before 1 March 2010). To avoid missing data, we also search more articles with keywords ‘acetylated’ and ‘acetyl’. After all substrates with unambiguous acetylation lysines were collected, we searched the UniProt Knowledgebase (23) to obtain the corresponding protein sequences and associated annotation information. The theoretical pI (isoelectric point) and Mw (molecular weight) were calculated for each protein (http://www.expasy.org/tools/pi_tool.html) (24,25).
In CPLA database, the PPI information was also integrated if available. We took experimental PPIs from several major public databases (on 10 April 2010), such as HPRD (20), BioGRID (26), DIP (27), MINT (28) and IntAct (29). The redundant PPIs were thoroughly cleared. In addition, a well-known pre-predicted database of STRING (30) was also used. All proteins were mapped to the UniProt sequences by BLAST. For human, we collected a total of 59 481 experimental PPIs in 12 221 proteins and 1 212 607 predicted PPIs in 16 523 proteins, respectively. The detailed statistics of PPI information was shown in Supplementary Table S1.
The CPLA 1.0 database contains 7151 lysine acetylation sites in 3311 substrates (Table 1). Particularly, 1742 (∼24.4%) acetylation sites in 726 proteins are collected from non-human species (Table 1). The online service and local packages were implemented in PHP + MySQL + JavaScript and JAVA 1.5 (J2SE 5.0), separately. Moreover, the online documentation and a user manual were also provided.
USAGE
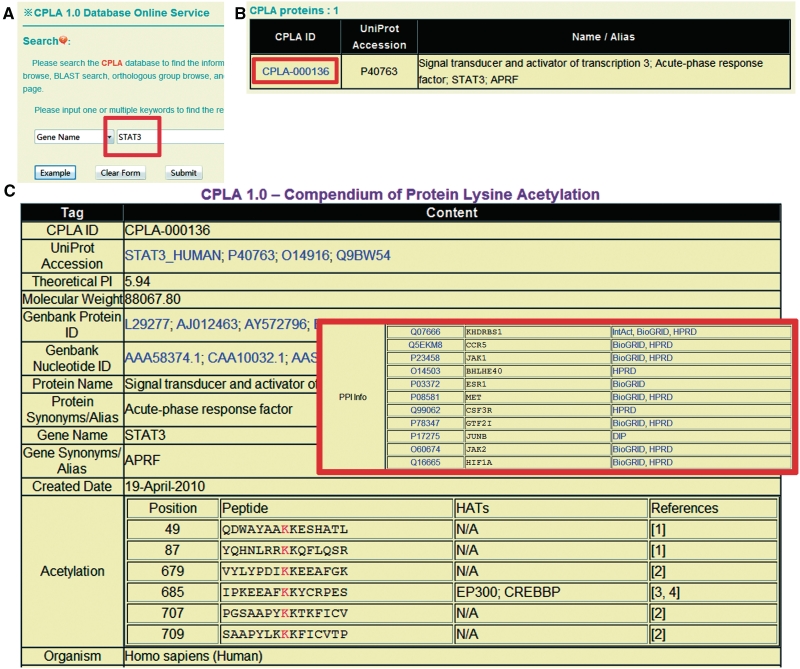
The CPLA database 1.0 was developed in a user-friendly manner. The search option (http://cpla.biocuckoo.org/search.php) provides an interface for querying the CPLA 1.0 database with one or several keywords such as gene/protein names, UniProt ID or CPLA ID, etc. For example, if the keyword ‘STAT3’ is inputted and submitted (Figure 1A), the result will be shown in a tabular format, with the features of CPLA ID, UniProt accession and protein/gene names/aliases (Figure 1B). By clicking on the CPLA ID (CPLA-000136), the detailed information for human STAT3 will be shown (Figure 1C). The acetylation information, including acetylated positions, flanking peptides, experimental reagents or upstream HATs, and primary references are provided. The protein sequence, GO annotation, domain organization, molecular weight, computed/ theoretical Ip and PPI information are also presented.
Figure 1.
The search option of CPLA 1.0 database. (A) Users could simply input ‘STAT3’ for querying. (B) The results will be shown in a tabular format. Users could click on the CPLA ID (CPLA-000025) to visualize the detailed information. (C) The detailed information of human STAT3. The PPI information was also presented.
Furthermore, we provided three additional advance options, including (i) advance search, (ii) browse and (iii) BLAST search (Supplementary Figure S1). (i) Advance search: in this option, users could use relatively complex and combined keywords to locate the precise information, with up to two search terms. The interface of search-engine permits the querying by different database fields and the linking of queries through three operators of ‘and’, ‘or’ and ‘exclude’ (Supplementary Figure S1A). (ii) Browse: instead of searching for a specific protein, all entries of CPLA database could be listed by species name (Supplementary Figure S1B). (iii) BLAST search: this option was designed for finding related information in CPLA database quickly. The blastall program of NCBI BLAST packages (31) was included in CPLA 1.0 database (Supplementary Figure S1C). Users can input a protein sequence in FASTA format for searching identical or homologous proteins.
RESULTS AND DISCUSSION
Recent progresses toward understanding the full functional content of acetylome have experimentally revealed several thousands of lysine acetylated substrates with their sites. Besides experimental efforts, computational studies such as predictor construction and database development also attract much attention. The current available computational resources were summarized and listed in Supplementary Table S2. Among these researches, database development is particularly important for integrating experimental data from heterogeneous sources, and providing a high quality benchmark for further experimental or computational designs. Although several public databases (19–22) have already maintained the acetylation information, the lysine acetylation is usually collected together with another less controlled Nα-terminal acetylation. In this work, we only focused on protein lysine acetylation and manually curated 7151 lysine acetylation sites in 3311 proteins.
Since a large proportion of acetylation sites were taken from Homo sapiens, we had the opportunity to analyze abundance and functional diversity of lysine acetylation in an acetylomic level. We surveyed the GO terms of 2585 acetylated proteins from UniProt annotations. Using the human proteome as the background, we statistically calculated over-represented biological processes, molecular functions and cellular components in acetylome with the hypergeometric distribution (P < 0.01). The top five most enriched GO entries in each category were shown in Table 2. Our analyses revealed several potentially interesting results. For example, the three most abundant biological processes such as translational elongation, RNA splicing and mRNA processing suggest that acetylation predominantly regulates gene expression in a post-transcriptional manner (Table 2). Also, four most over-represented molecular functions such as ATP binding, protein binding, RNA binding and nucleotide binding suggest that acetylation modulates enzyme activity and protein interaction ability (Table 2). In addition, the statistical analysis of cellular components revealed acetylated proteins to be highly enriched in distinct cellular compartments. For instance, ∼30 and ∼62% of cytosol and mitochondrial matrix proteins are acetylated, respectively (Table 2). For more detailed information, the top 15 most over-represented GO terms and InterPro domains were shown in Supplementary Tables S3 and S4.
Table 2.
The top five most enriched GO terms of biological processes, molecular functions and cellular components in human acetylome
| Description of GO term | Acetylome | Proteome | E-ratioc | P-value |
|---|---|---|---|---|
| n (%)a,b | n (%) | |||
| The top five most enriched biological processes | ||||
| Translational elongation (GO:0006414) | 69 (2.80) | 93 (0.52) | 5.44 | 3.35E-40 |
| Nucleosome assembly (GO:0006334) | 43 (1.75) | 79 (0.44) | 3.99 | 1.17E-17 |
| RNA splicing (GO:0008380) | 70 (2.84) | 188 (1.04) | 2.73 | 3.50E-16 |
| Interspecies interaction between organisms (GO:0044419) | 100 (4.06) | 333 (1.85) | 2.20 | 3.97E-15 |
| mRNA processing (GO:0006397) | 65 (2.64) | 193 (1.07) | 2.47 | 9.06E-13 |
| The top five most enriched molecular functions | ||||
| ATP binding (GO:0005524) | 398 (16.17) | 1473 (8.17) | 1.98 | 4.67E-46 |
| Protein binding (GO:0005515) | 798 (32.06) | 3996 (22.16) | 1.45 | 9.00E-35 |
| RNA binding (GO:0003723) | 186 (7.56) | 557 (3.09) | 2.45 | 9.49E-34 |
| Structural constituent of ribosome (GO:0003735) | 71 (2.89) | 155 (0.86) | 3.36 | 1.99E-22 |
| Nucleotide binding (GO:0000166) | 85 (3.45) | 255 (1.41) | 2.44 | 5.75E-16 |
| The top five most enriched cellular components | ||||
| Cytosol (GO:0005829) | 298 (12.11) | 1009 (5.60) | 2.16 | 4.71E-42 |
| Mitochondrial matrix (GO:0005759) | 83 (3.37) | 139 (0.77) | 4.38 | 6.33E-37 |
| Nucleoplasm (GO:0005654) | 124 (5.04) | 333 (1.85) | 2.73 | 1.12E-27 |
| Nucleolus (GO:0005730) | 133 (5.40) | 425 (2.36) | 2.29 | 1.84E-21 |
| Cytosolic small ribosomal subunit (GO:0022627) | 29 (1.18) | 36 (0.20) | 5.90 | 2.24E-19 |
aNum., number of proteins annotated; bPer. percentiles of proteins annotated; cE-ratio, enrichment ratio of acetylated proteins.
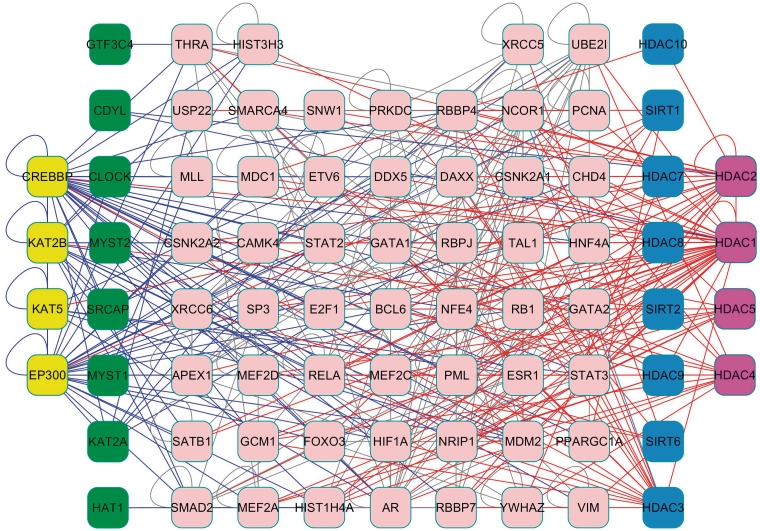
The acetylation and deacetylation of proteins are carried out by HATs and HDACs, which antagonistically and dynamically control protein function. Combined with experimental and predicted PPIs, we constructed a potential HLAN among HATs, substrates and HDACs, with 1019 PPIs of 199 proteins (Supplementary Table S5). If only experimental PPIs are considered, the core HLAN contained 369 PPIs among 77 proteins, including 12 HATs and 12 HDACs (Figure 2). From the whole HLAN, we retrieved 1862 potential triplet relations of HAT–substrate–HDAC (Supplementary Table S6). If a substrate is a HAT or HDAC, it should be acetylated or deacetylated by a different HAT or HDAC. We carefully surveyed scientific literature and found that at least 13 triplet interactions were experimentally identified (Supplementary Table S6). For example, Gaughan et al. (32) observed that Tip60 (KAT5) and histone deacetylase 1 (HDAC1) regulate the transcriptional activity of androgen receptor (AR) through changing its acetylation status, and form a KAT5-AR-HDAC1 relation (Supplementary Table S6). Moreover, our results also discovered a number of potentially interesting results. For instance, EP300 acetylates BCL6 at K379 and inhibits its function, while deacetylases were not clearly identified (33). In our results, the EP300-BCL6-HDAC5, EP300-BCL6-SIRT2, EP300-BCL6-HDAC11, EP300-BCL6-HDAC3, EP300-BCL6-HDAC2 and EP300-BCL6-HDAC8 suggested that BCL6 might be deacetylated by multiple HDACs (Supplementary Table S6). Moreover, human GCMa/GCM1 was reported to be acetylated by CBP/CREBBP at K367, K406 and K409 (34). In our results, the relations of CREBBP-GCM1-HDAC3, CREBBP-GCM1-HDAC3, CREBBP-GCM1-HDAC1 and CREBBP-GCM1-HDAC4 proposed that at least four HDACs might deacetylate GCM1 (Supplementary Table S6).
Figure 2.
A core HLAN identified from experimentally identified PPI data. The HATs, substrates, and HDACs form a dense network. Green node: HAT; Blue node: HDAC; yellow node: HAT that is acetylated; purple node: HDAC that is acetylated; pink node: substrate.
Taken together, here we developed a comprehensive database of protein lysine acetylation. The statistical analyses revealed functional diversity and enrichment of acetylation, while network studies generated a large number of potentially useful results for further experimental or computational researches. The CPLA database will be routinely updated if new acetylated substrates are reported.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Funding for open access charge: National Basic Research Program (973 project) (2010CB945400, 2007CB947401); National Natural Science Foundation of China (90919001, 30700138, 30900835, 30830036, 31071154); Chinese Academy of Sciences (INFO-115-C01-SDB4-36).
Conflict of interest statement. None declared.
REFERENCES
- 1.Polevoda B, Sherman F. Nalpha -terminal acetylation of eukaryotic proteins. J. Biol. Chem. 2000;275:36479–36482. doi: 10.1074/jbc.R000023200. [DOI] [PubMed] [Google Scholar]
- 2.Polevoda B, Sherman F. The diversity of acetylated proteins. Genome Biol. 2002;3:reviews0006. doi: 10.1186/gb-2002-3-5-reviews0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Smith KT, Workman JL. Introducing the acetylome. Nat. Biotechnol. 2009;27:917–919. doi: 10.1038/nbt1009-917. [DOI] [PubMed] [Google Scholar]
- 4.Yang XJ, Seto E. The Rpd3/Hda1 family of lysine deacetylases: from bacteria and yeast to mice and men. Nat. Rev. Mol. Cell. Biol. 2008;9:206–218. doi: 10.1038/nrm2346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yang XJ, Seto E. HATs and HDACs: from structure, function and regulation to novel strategies for therapy and prevention. Oncogene. 2007;26:5310–5318. doi: 10.1038/sj.onc.1210599. [DOI] [PubMed] [Google Scholar]
- 6.Lee KK, Workman JL. Histone acetyltransferase complexes: one size doesn't fit all. Nat. Rev. Mol. Cell Biol. 2007;8:284–295. doi: 10.1038/nrm2145. [DOI] [PubMed] [Google Scholar]
- 7.Yang XJ. The diverse superfamily of lysine acetyltransferases and their roles in leukemia and other diseases. Nucleic Acids Res. 2004;32:959–976. doi: 10.1093/nar/gkh252. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kouzarides T. Acetylation: a regulatory modification to rival phosphorylation? EMBO J. 2000;19:1176–1179. doi: 10.1093/emboj/19.6.1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Allfrey VG, Faulkner R, Mirsky AE. Acetylation and methylation of histones and their possible role in the regulation of Rna synthesis. Proc. Natl Acad. Sci. USA. 1964;51:786–794. doi: 10.1073/pnas.51.5.786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yuan ZL, Guan YJ, Chatterjee D, Chin YE. Stat3 dimerization regulated by reversible acetylation of a single lysine residue. Science. 2005;307:269–273. doi: 10.1126/science.1105166. [DOI] [PubMed] [Google Scholar]
- 11.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, Olsen JV, Mann M. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009;325:834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 12.Walkinshaw DR, Tahmasebi S, Bertos NR, Yang XJ. Histone deacetylases as transducers and targets of nuclear signaling. J. Cell. Biochem. 2008;104:1541–1552. doi: 10.1002/jcb.21746. [DOI] [PubMed] [Google Scholar]
- 13.Brunet A, Sweeney LB, Sturgill JF, Chua KF, Greer PL, Lin Y, Tran H, Ross SE, Mostoslavsky R, Cohen HY, et al. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303:2011–2015. doi: 10.1126/science.1094637. [DOI] [PubMed] [Google Scholar]
- 14.Kim SC, Sprung R, Chen Y, Xu Y, Ball H, Pei J, Cheng T, Kho Y, Xiao H, Xiao L, et al. Substrate and functional diversity of lysine acetylation revealed by a proteomics survey. Mol. Cell. 2006;23:607–618. doi: 10.1016/j.molcel.2006.06.026. [DOI] [PubMed] [Google Scholar]
- 15.Wang Q, Zhang Y, Yang C, Xiong H, Lin Y, Yao J, Li H, Xie L, Zhao W, Yao Y, et al. Acetylation of metabolic enzymes coordinates carbon source utilization and metabolic flux. Science. 2010;327:1004–1007. doi: 10.1126/science.1179687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zhao S, Xu W, Jiang W, Yu W, Lin Y, Zhang T, Yao J, Zhou L, Zeng Y, Li H, et al. Regulation of cellular metabolism by protein lysine acetylation. Science. 2010;327:1000–1004. doi: 10.1126/science.1179689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cohen HY, Miller C, Bitterman KJ, Wall NR, Hekking B, Kessler B, Howitz KT, Gorospe M, de Cabo R, Sinclair DA. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 18.Zhang J, Sprung R, Pei J, Tan X, Kim S, Zhu H, Liu CF, Grishin NV, Zhao Y. Lysine acetylation is a highly abundant and evolutionarily conserved modification in Escherichia coli. Mol. Cell. Proteomics. 2009;8:215–225. doi: 10.1074/mcp.M800187-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hornbeck PV, Chabra I, Kornhauser JM, Skrzypek E, Zhang B. PhosphoSite: a bioinformatics resource dedicated to physiological protein phosphorylation. Proteomics. 2004;4:1551–1561. doi: 10.1002/pmic.200300772. [DOI] [PubMed] [Google Scholar]
- 20.Keshava Prasad TS, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicherla D, Raju R, Shafreen B, Venugopal A, et al. Human protein reference database–2009 update. Nucleic Acids Res. 2009;37:D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li H, Xing X, Ding G, Li Q, Wang C, Xie L, Zeng R, Li Y. SysPTM - a systematic resource for proteomic research of post-translational modifications. Mol Cell Proteomics. 2009;8:1839–1849. doi: 10.1074/mcp.M900030-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lee TY, Hsu JB, Chang WC, Wang TY, Hsu PC, Huang HD. A comprehensive resource for integrating and displaying protein post-translational modifications. BMC Res. Notes. 2009;2:111. doi: 10.1186/1756-0500-2-111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.(2010) The Universal Protein Resource. (UniProt) in 2010. Nucleic Acids Res. 38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Bjellqvist B, Basse B, Olsen E, Celis JE. Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis. 1994;15:529–539. doi: 10.1002/elps.1150150171. [DOI] [PubMed] [Google Scholar]
- 25.Bjellqvist B, Hughes GJ, Pasquali C, Paquet N, Ravier F, Sanchez JC, Frutiger S, Hochstrasser D. The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis. 1993;14:1023–1031. doi: 10.1002/elps.11501401163. [DOI] [PubMed] [Google Scholar]
- 26.Stark C, Breitkreutz BJ, Reguly T, Boucher L, Breitkreutz A, Tyers M. BioGRID: a general repository for interaction datasets. Nucleic Acids Res. 2006;34:D535–D539. doi: 10.1093/nar/gkj109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Salwinski L, Miller CS, Smith AJ, Pettit FK, Bowie JU, Eisenberg D. The database of interacting proteins: 2004 update. Nucleic Acids Res. 2004;32:D449–D451. doi: 10.1093/nar/gkh086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chatr-aryamontri A, Ceol A, Palazzi LM, Nardelli G, Schneider MV, Castagnoli L, Cesareni G. MINT: the Molecular INTeraction database. Nucleic Acids Res. 2007;35:D572–D574. doi: 10.1093/nar/gkl950. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Aranda B, Achuthan P, Alam-Faruque Y, Armean I, Bridge A, Derow C, Feuermann M, Ghanbarian AT, Kerrien S, Khadake J, et al. The IntAct molecular interaction database in 2010. Nucleic Acids Res. 2010;38:D525–D531. doi: 10.1093/nar/gkp878. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Jensen LJ, Kuhn M, Stark M, Chaffron S, Creevey C, Muller J, Doerks T, Julien P, Roth A, Simonovic M, et al. STRING 8–a global view on proteins and their functional interactions in 630 organisms. Nucleic Acids Res. 2009;37:D412–D416. doi: 10.1093/nar/gkn760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Johnson M, Zaretskaya I, Raytselis Y, Merezhuk Y, McGinnis S, Madden TL. NCBI BLAST: a better web interface. Nucleic Acids Res. 2008;36:W5–W9. doi: 10.1093/nar/gkn201. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gaughan L, Logan IR, Cook S, Neal DE, Robson CN. Tip60 and histone deacetylase 1 regulate androgen receptor activity through changes to the acetylation status of the receptor. J. Biol. Chem. 2002;277:25904–25913. doi: 10.1074/jbc.M203423200. [DOI] [PubMed] [Google Scholar]
- 33.Bereshchenko OR, Gu W, Dalla-Favera R. Acetylation inactivates the transcriptional repressor BCL6. Nat. Genet. 2002;32:606–613. doi: 10.1038/ng1018. [DOI] [PubMed] [Google Scholar]
- 34.Chang CW, Chuang HC, Yu C, Yao TP, Chen H. Stimulation of GCMa transcriptional activity by cyclic AMP/protein kinase A signaling is attributed to CBP-mediated acetylation of GCMa. Mol. Cell. Biol. 2005;25:8401–8414. doi: 10.1128/MCB.25.19.8401-8414.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]