Abstract
KUPS (The University of Kansas Proteomics Service) provides high-quality protein–protein interaction (PPI) data for researchers developing and evaluating computational models for predicting PPIs by allowing users to construct ready-to-use data sets of interacting protein pairs (IPPs), non-interacting protein pairs (NIPs) and associated features. Multiple filters and options allow the user to control the make-up of the IPPs and NIPs as well as the quality of the resultant data sets. Each data set is built from the overall database, which includes 185 446 IPPs and ∼1.5 billion NIPs from five primary databases: IntAct, HPRD, MINT, UniProt and the Gene Ontology. The IPP set can be set to specific model organisms, interaction types and experimental evidence. The NIP set can be generated using four different strategies, which can alleviate biased estimation problems. Lastly, multiple features can be provided for all of the IPP and NIP pairs. Additionally, KUPS provides two benchmark data sets to help researchers compare their algorithms to existing approaches. KUPS is freely available at http://www.ittc.ku.edu/chenlab.
INTRODUCTION
Proteins are the fundamental units of life, which play significant roles in cellular processes such as composing cellular structure and promoting chemical reactions. The multiplicity of functions that proteins execute in most cellular processes and biochemical events is attributed to their interactions with other proteins. Thus, understanding protein–protein interactions (PPIs) will help to elucidate cellular processes and functions, identify pharmacological targets and design drugs.
While advanced high-throughput technologies are providing a large amount of new PPI data, the number of discovered PPIs is still far from complete. Therefore, there is a critical need to build in silico models that can predict PPIs existing in vivo. To develop and evaluate computational models for PPI prediction, two essential components are required: (i) positive data (interacting protein pairs or IPP) and negative data (non-interacting protein pairs or NIP) and (ii) features (attributes) for each data point (instance). While IPP data can be downloaded from some existing databases, generating NIP data is not a trivial task. Furthermore, there is no existing database that provides users with the second component, the features.
Negatome (1) is the first database that provides users with experimentally supported mammalian NIPs. These NIPs are derived from manually curated literature and protein complexes. Negatome is limited in developing PPI prediction algorithms because of the lack of IPPs and insufficient number of NIPs. To build a reliable PPI prediction model with generalization capabilities, the number of negatives is expected to be several orders of magnitude higher than the number of positives (2,3). For example, in yeast, ∼6000 proteins allow for ∼18 million potential interacting pairs; among them, <100 000 pairs are estimated to form actual interactions (2,4–6). With <2000 NIPs and zero IPPs, negatome data cannot be used as a sole source for training PPI prediction algorithms.
GRIP (7) is a web-based system that provides both IPPs and NIPs in Saccharomyces cerevisiae. In the GRIP, IPPs are extracted from the MIPS Comprehensive Yeast Genome Database (8), which includes only proteins in the same complex. NIPs are created by using uniform random sampling of proteins belonging to different subcellular locations. Although both IPPs and NIPs are available for users, GRIP database is limited in that the data are extracted from a single species (yeast) and are biased due to the mechanisms used in negative data generation [studies show that choosing negative data in terms of different subcellular localizations leads to biased estimates of system performance (9)].
Another limitation existing in both the negatome and GRIP is that they both simply provide a list of protein pairs without features or attributions for each example. In algorithm development, feature extraction is one of the most important components that can significantly affect the performance of computational models. To facilitate the development and evaluation of new computational methods for PPI prediction, we develop KUPS (The University of Kansas Proteome Service) database, which addresses the existing issues by providing a significantly enlarged and enriched user-friendly database. Compared with the negatome and GRIP, KUPS allows users to create databases with larger scale (in terms of the number of IPPs and NIPs) and larger coverage (in terms of species involved). Furthermore, KUPS offers new options that are not available in both the negatome and GRIP: (i) users can construct ready-to-use data sets including both IPPs and NIPs with the associated features, rather than a simple list of protein pairs; (ii) users can construct data sets with examples of various levels of quality (e.g. high-throughput experiments, biochemical methods or inference) and different types (e.g. direct interaction and enzymatic reaction); and (iii) users can construct NIPs using four different strategies (discussed next) to alleviate the biased estimation problems. In addition, KUPS creates two benchmark data sets for researchers to develop and test their algorithms: one data set with balanced IPPs and NIPs and the other with imbalanced IPPs and NIPs, which reflects the observed sparsity of protein interactions. Table 1 summarizes the three databases. The KUPS is freely available through our website (http://www.ittc.ku.edu/chenlab/).
Table 1.
Comparing three databases
| Databases |
|||
|---|---|---|---|
| Functions | Negatome | GRIP | KUPS |
| Users create IPPs? | No | Yes | Yes |
| Maximum no. of possible IPPs | 0 | 10 994 | 185 446 |
| Maximum no. of possible NIPs | 1892 | 319 855 | 1.5 billion |
| Un-biased NIPs? | Yes | No | Yes |
| No. of model organisms | NAa | 1 | 8 |
| Choice of data quality and interaction types | No | No | Yes |
| Methods to create NIPs | Literature curation and structural information | Sub-cellular localization | Four strategies |
| Benchmarks | No | No | Yes |
| Feature extraction | No | No | Yes |
| Ready-to-use? | No | No | Yes |
aNegative PPIs obtained from literature curation are extracted from mammalian proteins (most from human data); and negative PPIs obtained from PDB are extracted from mammalians (47%) and other species (53%).
THE DATABASE
Overview
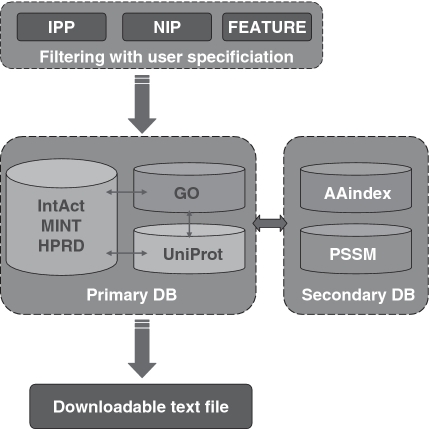
KUPS is built by integrating five databases [MINT (10), IntAct (11), HPRD (12), Gene Ontology (GO) (13) and UniProt (14)] into a primary database and two other databases [AAindex (15) and PSSM (16)] into a secondary database. Powerful filtering interfaces are provided to effectively handle the information from multiple databases and users. The structure of KUPS is shown in Figure 1.
Figure 1.
Structure of KUPS.
Through front-end interfaces, users can specify three different groups of filters to effectively control the distributions and quality of positive (IPP) and negative (NIP) interacting protein pairs and the types of features (FEATURE) to use in the final output.
The primary database controls the population and distributions of examples. It integrates five independent databases: the three PPI databases (IntAct, MINT and HPRD) are used for extracting and generating IPPs and NIPs; GO is used for calculating the distance of annotations for generating NIPs and UniProt is used for referring annotations of individual proteins and their sequences. AAindex and PSSM are considered the secondary database in KUPS because they are not directly involved to define the list of final data set. However, they are used for generating features of the final PPI data. Finally, the generated IPP/NIP data set is written into a text file downloadable by users.
In addition, KUPS provides a data repository that includes benchmark data sets with evaluation results of commonly used learning models and testing data sets used in other studies.
Integrated databases
Five databases are integrated into KUPS as the primary database to generate both IPPs and NIPs. IPPs are extracted from three PPI databases (IntAct, MINT and HPRD) downloaded with a PSI-MI format. Essential information, such as species, gene name, interaction type and detection method, is extracted using our own parsing program. During the parsing process, a pair of interacting proteins is omitted if any one of the required information (i.e. protein names, interaction types and detection methods) is missed. Only unique pairs are stored in KUPS by avoiding redundancy among the three databases. After parsing, KUPS has stored ∼185 446 PPI pairs from various organisms (the distribution for eight main model organisms is shown in Figure 2).
Figure 2.
KUPS PPI distributions for model organisms.
GO database consists of three categories of a gene product: molecular function, biological process and cellular component. The terms in the GO database are used to annotate database entries like UniProt to describe the roles of genes and gene products in any organism. GO also provides ‘Evidence Codes’ that are controlled vocabulary describing the nature of evidence to support a particular association. UniProt provides manually curated stable, comprehensive, freely accessible central resource on protein sequences and functional annotation and is often used as cross-references to other databases. UniProt is essential in KUPS by providing the links between PPI databases and GO database because some PPI databases do not provide annotations or related protein sequences.
KUPS provides not only IPPs and NIPs but also enriched information about each protein such as protein sequence, PSSM and converted real value features. This additional information of each unique protein is gathering from secondary database through the request of external users. In the secondary database, AAindex provides numerical scales of physicochemical and biochemical properties of amino acids by manually curating amino acid scales from research papers. Many studies (17–20) have used physicochemical and biochemical properties to elucidate cellular functions or processes; therefore, KUPS adopts AAindex to define features, which can be used for user’s own purpose on designing in silico methods for PPI predictions. Another database, PSSM, was first introduced for detecting distantly related proteins based on the probability of amino acid appearance at each position by using previously aligned group of protein sequences or structures. Generating PSSMs is time consuming and normally requires a high performance machine and background knowledge about operating the software and machine. Therefore, KUPS provides PSSMs for each protein in the list of final data set by using PSI-BLAST (21). To create PSSM profiles in KUPS, the e-value and number of iterations are set to be 0.001 and 3, respectively. We use the default values set by PSI-BLAST for other parameters.
Filters with users’ specification
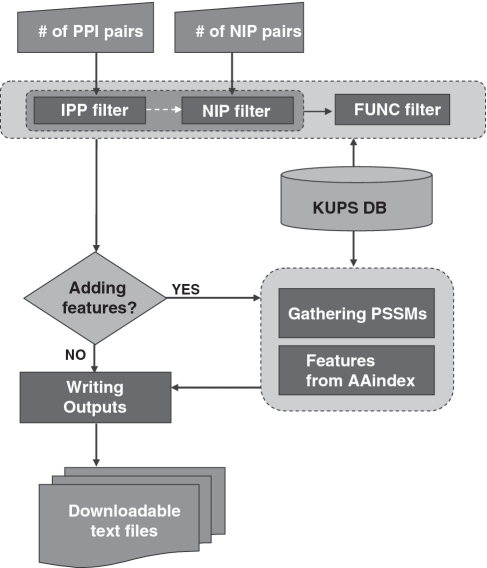
KUPS provides various filtering processes for users to specify their needs. Figure 3 shows the workflow diagram with three filters: IPP filter for specifying positive interacting protein pairs; NIP filter for specifying non-interacting protein pairs and FUNC filter for specifying features to extract.
Figure 3.
Workflow diagram in KUPS.
There are four adjustable parameters in IPP filter: number of IPPs, species, interaction types and detection method. Except for the number of IPPs, all other parameters have multiple choices and by default all choices for each parameter are used. In KUPs, IPPs can be extracted from 13 species with different interaction types (e.g. physical association) and detection methods (e.g. biochemical methods or two hybrid methods). After users choose the parameters, KUPS searches the entire database and returns PPI pairs that match the filtering parameters.
The NIP filter controls the populations and distributions of non-interacting protein pairs. Besides the number of NIPs to generate, it has five more parameters that allow for producing eight different negative sets. By checking the first parameter ‘Restrict protein selection pool’, the NIPs are restricted to the combinations of proteins appeared in the previously generated IPPs. If this parameter is unchecked, then the entire database is used for generating NIPs. Four different methods are used for creating NIPs: uniform random pairs; functionally dissimilar pairs; spatially separated pairs and non-interacting domains.
Uniform random pairs are often considered less biased than molecular processes-based selection methods. They create NIPs by randomly selecting possible interacting pairs that are not appeared in known PPI lists.
Functionally dissimilar pairs are also known to be less biased than spatially separated NIP selection methods (9). KUPS first calculates semantic similarity between two proteins according to Lord et al. (22,23). The assumption is that the most dissimilar annotation pair will have the smallest similarity score. Potential non-interacting protein pairs are sorted by their similarity scores.
Spatially separate pairs define NIPs based on annotations of cellular component by selecting protein pairs that do not have any overlapping cellular component annotations.
Non-interacting domain pairs create NIPs based on non-interacting domain pairs defined in the negatome database (1): all proteins in KUPS are first mapped into Pfam (24) to identify their domains; KUPS selects protein pairs with non-interacting domains as NIPs.
Features generation
Besides creating the list of interacting and non-interacting protein pairs, KUPS provides eight groups of features for algorithm design and evaluation. This is an important component that allows users to focus on designing and evaluating computational algorithms for PPI predictions. The first six feature sets are ‘Interaction type’, ‘Detection method’, ‘Species’, ‘Locality’ and ‘GO annotation’ that describe the data users generate. The other three groups of features are extracted from protein sequences as described next.
Studies have shown that amino acid sequences contain significant information for characterizing protein interactions (25–31). However, extracting features from protein sequences is often time consuming and requires large computational power. KUPS provides several sequence-based features for users to evaluate. The first group of features is amino acid sequences for each protein. The second feature set is AAindex-based sequence features (15), which include the scales of physicochemical or biochemical properties. PSSM profile is another set of features that gives the log-odds score of each amino acid in each position of a target protein sequence. It has shown great potential in protein secondary structure prediction (32) and function prediction (33). KUPS provides PSSM profiles created by PSI-BLAST for each protein.
With KUPS, users can choose all or combinations of these eight groups of features and create a data set of IPPs and NIPs for algorithm development.
TEST SET COLLECTION AND BENCHMARK
KUPS has a collection of test sets used in research publications on PPI inference for researchers intending to compare their algorithms to other existing methods. The test set collection has three categories: methods for predicting PPIs, predicting protein functions and predicting protein interface residues. Each category has information about the published papers and downloadable data sets. Data sets are collected directly from authors’ websites or Supplementary Materials of the paper.
Two benchmark data sets generated by this service are also included for comparing algorithm performance to existing solutions. The first is balanced with equal interacting and non-interacting protein pairs in both the training and test sets. The second is an imbalanced data set, with significantly more non-interacting pairs than interacting pairs. For machine-learning approaches, correct classification is easier in balanced data sets; however, the imbalanced data set more closely reflects the observed sparsity of interacting protein pairs. The performance of learning models on the benchmarks is presented with seven common measurements: overall accuracy, specificity, recall, precision, f-measure, correlation coefficient and confusion matrix. Results from four common learning models (Naïve Bayes, decision tree, support vector machine and random forests) are already available. Researchers are encouraged to share their algorithm’s results to the benchmarks for inclusion on the website. The goal is to provide as complete a comparison between existing algorithms as possible.
CONCLUSIONS
To the best of our knowledge, KUPS is the first database that provides ready-to-use data sets for researchers to use. Compared with the other two existing databases, KUPS offers larger coverage (number of organisms where PPIs are derived) and larger scale (total number of PPIs). It allows users not only to generate positive and negative PPI data but also to create associated features as well, while other databases often provide users with a list of positive and negative PPI pair names only. KUPS allows for a fair comparison and evaluation of different learning methods for PPI prediction, as different methods can be applied to the same data sets with the same features generated from KUPS. In addition, users can generate a highly imbalanced data set with significantly more negative examples than positive examples, which mirrors the observed sparsity of true protein interactions. Furthermore, users can generate negative examples with KUPS using four different strategies to avoid the bias problems in GRIP.
FUNDING
National Science Foundation (IIS-0644366).
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Matthaw Mandelbaum who has provided valuable discussions.
REFERENCES
- 1.Smialowski P, Pagel P, Wong P, Brauner B, Dunger I, Fobo G, Frishman G, Montrone C, Rattei T, Frishman D, et al. The Negatome database: a reference set of non-interacting protein pairs. Nucleic Acids Res. 2009;38(Database issue):D540–D544. doi: 10.1093/nar/gkp1026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jansen R, Yu H, Greenbaum D, Kluger Y, Krogan N, Ching S, Emili A, Snyder M, Greenblatt J, Gerstein M. A Bayesian networks approach for predicting protein-protein interactions from genomic data. Science. 2003;302:449–453. doi: 10.1126/science.1087361. [DOI] [PubMed] [Google Scholar]
- 3.Jansen R, Gerstein M. Analyzing protein function on a genomic scale: the importance of gold-standard positives and negatives for network prediction. Curr. Opin. Microbiol. 2004;7:535–545. doi: 10.1016/j.mib.2004.08.012. [DOI] [PubMed] [Google Scholar]
- 4.Bader G, Hogue C. Analyzing yeast protein-protein interaction data obtained from different sources. Nat. Biotechnol. 2002;20:991–997. doi: 10.1038/nbt1002-991. [DOI] [PubMed] [Google Scholar]
- 5.Grigoriev A. On the number of protein-protein interactions in the yeast proteome. Nucleic Acids Res. 2003;31:4157–4161. doi: 10.1093/nar/gkg466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.von Mering C, Krause R, Snel B, Cornell M, Oliver S, Fields S, Bork P. Comparative assessment of large-scale data sets of protein-protein interactions. Nature. 2002;417:399–403. doi: 10.1038/nature750. [DOI] [PubMed] [Google Scholar]
- 7.Browne F, Wang H, Zheng H, Azuaje F. GRIP: A web-based system for constructing gold standard datasets for protein-protein interaction prediction. Source Code Biol. Med. 2009;4:2. doi: 10.1186/1751-0473-4-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mewes H, Albermann K, Heumann K, Liebl S, Pfeiffer F. MIPS: a database for protein sequences, homology data and yeast genome information. Nucleic Acids Res. 1997;25:28–30. doi: 10.1093/nar/25.1.28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Ben-Hur A, Noble W. Choosing negative examples for the prediction of protein-protein interactions. BMC Bioinformaticss. 2006;7(Suppl. 1):S1. doi: 10.1186/1471-2105-7-S1-S2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Ceol A, Aryamontri A, Licata L, Peluso D, Briganti L, Perfetto L, Castagnoli L, Cesareni G. MINT, the molecular interaction database: 2009 update. Nucleic Acids Res. 2010;38(Database issue):D532–D539. doi: 10.1093/nar/gkp983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hermjakob H, Montecchi-Palazzi L, Lewington C, Mudali S, Kerrien S, Orchard S, Vingron M, Roechert B, Roepstorff P, Valencia A. IntAct: an open source molecular interaction database. Nucleic Acids Res. 2004;32(Database issue):D452–D455. doi: 10.1093/nar/gkh052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Keshava Prasad T, Goel R, Kandasamy K, Keerthikumar S, Kumar S, Mathivanan S, Telikicheria D, Raju R, Shafreen B, Venugopal A. Human protein reference database0—2009 update. Nucleic Acids Res. 2009;37(Database issue):D767–D772. doi: 10.1093/nar/gkn892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.The Gene Ontology Consortium. The Gene ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38(Suppl. 1):D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.The UniProt Consortium. The universal protein resource (UniProt)in 2010. Nucleic Acids Res. 2010;38(Suppl. 1):D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kawashima S, Ogata H, Kanehisa M. AAindex: amino acid index database. Nucleic Acids Res. 1999;27:368–369. doi: 10.1093/nar/27.1.368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gribskov M, McLachlan A, Eisenberg D. Profile analysis: detection of distantly related proteins. Proc. Natl Acad. Sci. USA. 1987;84:4355–4358. doi: 10.1073/pnas.84.13.4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Garian R. Prediction of quaternary structure from primary structure. Bioinformatics. 2001;17:551–556. doi: 10.1093/bioinformatics/17.6.551. [DOI] [PubMed] [Google Scholar]
- 18.Garg A, Raghava G. A machine learning based method for the prediction of secretory proteins using amino acid composition, their order and similarity-search. In Silico Biol. 2008;8:129–140. [PubMed] [Google Scholar]
- 19.Bhasin M, Raghava G. Analysis and prediction of affinity of TAP binding peptides using cascade SVM. Protein Sci. 2004;13:596–607. doi: 10.1110/ps.03373104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen X, Jeong J. Sequence-based prediction of protein interaction sites with an integrative method. Bioinformatics. 2009;25:585–591. doi: 10.1093/bioinformatics/btp039. [DOI] [PubMed] [Google Scholar]
- 21.Altschul S, Madden T, Schaffer A, Zhang J, Zhang Z, Miller W, Lipman D. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lord P, Stevens R, Brass A, Goble A. Investigating semantic similarity measures across the Gene Ontology: the relationship between sequence and annotation. Bioinformatics. 2003;19:1275–1283. doi: 10.1093/bioinformatics/btg153. [DOI] [PubMed] [Google Scholar]
- 23.Lord P, Stevens R, Brass A, Goble A. Semantic similarity measures as tools for exploring the gene ontology. Pac. Symp. Biocomput. 2003;8:601–612. doi: 10.1142/9789812776303_0056. [DOI] [PubMed] [Google Scholar]
- 24.Finn R, Mistry J, Tate J, Coggill P, Heger A, Pollington J, Gavin L, Gunasekaran P, Ceric G, Forslund K. The Pfam protein families database. Nucleic Acids Res. 2010;38(Suppl. 1):D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bock J, Gough D. Predicting protein–protein interactions from primary structure. Bioinformatics. 2001;17:455–460. doi: 10.1093/bioinformatics/17.5.455. [DOI] [PubMed] [Google Scholar]
- 26.Hopp T, Woods K. Prediction of protein antigenic determinants from amino acid sequences. Proc. Natl Acad. Sci. USA. 1981;78:3824–3828. doi: 10.1073/pnas.78.6.3824. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Guo Y, Yu L, Wen Z, Li M. Using support vector machine combined with auto covariance to predict protein-protein interactions from protein sequences. Nucleic Acids Res. 2008;36:3025–3030. doi: 10.1093/nar/gkn159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jones S, Thornton J. Principles of protein-protein interactions. Proc. Natl Acad. Sci. USA. 1996;93:13–20. doi: 10.1073/pnas.93.1.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones S, Thornton J. Prediction of protein-protein interaction sites using patch analysis. J. Mol. Biol. 1997;272:133–143. doi: 10.1006/jmbi.1997.1233. [DOI] [PubMed] [Google Scholar]
- 30.Jones S, Thornton J. Analysis of protein-protein interaction sites using surface patches. J. Mol. Biol. 1997;272:121–132. doi: 10.1006/jmbi.1997.1234. [DOI] [PubMed] [Google Scholar]
- 31.Shen J, Zhang J, Luo X, Zhu W, Yu K, Chen K, Li Y, Jiang H. Predicting protein-protein interactions based only on sequences information. Proc. Natl Acad. Sci. USA. 2007;104:4337–4341. doi: 10.1073/pnas.0607879104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jones D. Protein secondary structure prediction based on position-specific scoring matrices. J. Mol. Biol. 1999;292:195–202. doi: 10.1006/jmbi.1999.3091. [DOI] [PubMed] [Google Scholar]
- 33.Jeong J, Lin X, Chen X. On position-specific scoring matrix for protein function prediciton. IEEE/ACM Trans. Comput. Biol. Bioinform. 2010 doi: 10.1109/TCBB.2010.93. http://doi.ieeecomputersociety.org/10.1109/TCBB.2010.93. [DOI] [PubMed] [Google Scholar]