Abstract
Numerous genetic variations have been found to be related to human diseases. Significant portion of those affect the drug response as well by changing the protein structure and function. Therefore, it is crucial to understand the trilateral relationship among genomic variations, diseases and drugs. We present the variations and drugs (VnD), a consolidated database containing information on diseases, related genes and genetic variations, protein structures and drug information. VnD was built in three steps. First, we integrated various resources systematically to deduce catalogs of disease-related genes, single nucleotide polymorphisms (SNPs), protein mutations and relevant drugs. VnD contains 137 195 disease-related gene records (13 940 distinct genes) and 16 586 genetic variation records (1790 distinct variations). Next, we carried out structure modeling and docking simulation for wild-type and mutant proteins to examine the structural and functional consequences of non-synonymous SNPs in the drug-related genes. Conformational changes in 590 wild-type and 4437 mutant proteins from drug-related genes were included in our database. Finally, we investigated the structural and biochemical properties relevant to drug binding such as the distribution of SNPs in proximal protein pockets, thermo-chemical stability, interactions with drugs and physico-chemical properties. The VnD database, available at http://vnd.kobic.re.kr:8080/VnD/ or vandd.org, would be a useful platform for researchers studying the underlying mechanism for association among genetic variations, diseases and drugs.
INTRODUCTION
Discovering genetic factors affecting disorders or diseases is crucial for understanding the pathogenesis, diagnosis and treatment of human diseases. Previous studies indicate that single nucleotide polymorphisms (SNPs) are the most common type of DNA sequence variation found in human genome, accounting for at least 1% of the genetic differences between individuals (1,2). In particular, non-synonymous SNPs (nsSNPs) in the coding region of a gene can alter the function or structure of protein by changing amino acids or introducing a premature stop codon (3). Conformational changes in these proteins are major targets for drug development. Indeed, drug response to these genetic variations has emerged to be a major subject in the field of pharmacogenomics with the combined use of genetics and functional genomics data. Information on SNPs and structural changes in disease-related proteins is thus important in biomedical studies, diagnostics and drug development (4).
Both public and commercial databases exist to provide information on relationship between genetic variants and drug targets. Such public efforts are represented by GenoWatch (5), IDBD (6), DrugBank (7) and SuperDrug (8). The GenoWatch and IDBD databases contain information about specific diseases and a browser for disease–gene association studies. DrugBank contains details on drugs such as drug target and action, and SuperDrug provides three-dimensional (3D) structures and conformers of drugs. Although each database has its own objectives, they provide information of limited scope such as disease-associated genes, genetic variations and drugs or 3D structural models of drugs. The commercial sector, led by the World Drug Index (9), Chemistry, Manufacturing and Controls (CMC) (10) and the MDL Drug Data Report (11), provides more comprehensive coverage. However, they are usually very expensive and accessible only by private commercial entities.
Protein structure modeling and docking simulations require computational power and experts. To our knowledge, no public resource is available to cover the structural aspect of disease proteins taking their genetic variations into account. Furthermore, effect of genetic variations on docking with drugs would be valuable information for drug development.
Here, we present a database, variations and drugs (VnD), which provides comprehensive information on diseases-related genes, their genetic variations, protein structure modeling and docking simulations. More specifically, available information is as follows: (i) a comprehensive catalog of disease-related genes, proteins and drugs; (ii) structural changes caused by nsSNPs in disease-related genes; (iii) their consequences in drug binding using docking simulation such as AutoDock (12), Dock (13) and Fred (14) programs; (iv) distribution of nsSNPs near the structural pockets in disease-related proteins; and (v) functional effects of SNPs known to be related to common diseases from association studies.
DATABASE DESIGN AND CONTENT
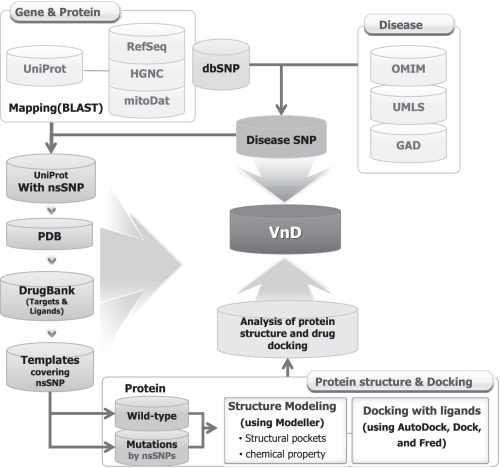
To build the VnD database, we developed an automatic pipeline as shown in Figure 1. It consists of three main steps: (i) collection of disease-related genetic variations and proteins from public disease databases using ontology-based unification of disease terms, (ii) structure modeling for both wild-type and nsSNP mutant proteins and (iii) analysis of protein structures and identification of potential drug binding sites.
Figure 1.
The workflow of the VnD database.
Collection of genetic variations associated with diseases
Disease term unification
We extracted disease terms from two disease databases: OMIM (15) and GAD (16). Unfortunately, these databases use highly inconsistent terminology to describe the same disease. For example, 141 slightly different disease descriptions exist for ‘Parkinson’s disease’. Therefore, we used the Unified Medical Language System (UMLS) (17), which contains medical subject headings (MeSHs) and clinical terms from the systematized nomenclature of medicine to standardize the disease terms. The disease terms in OMIM and GAD were mapped on the concept unique identifier (CUI) in UMLS (18) taking disease synonyms into consideration. Through this unification procedure, we obtained 36 109 disease terms, which were then mapped to 3898 CUIs (see Supplementary Table S1 for statistics).
Collection of disease-related genes
We extracted the candidate genes associated with diseases or disorders based on genomic positions and gene names. To cover the name space of disease-related genes, we extracted 40 234 gene names from the HUGO Gene Nomenclature Committee (HGNC) (19) and the NCBI Gene database (20). We integrated the genome annotation data as well from various sources: NCBI’s Entrez Gene (20), RefSeq mRNA from the UCSC table track (21) and protein information from UniProt (22). RefSeq mRNAs were mapped to genes, and 85 510 proteins were linked to genes using the BLAST (23) search. Ultimately, we obtained 13 940 disease-related genes and 10 883 disease-related UniProt proteins (Supplementary Table S2).
Collection of disease-related genetic variations
As a source of genetic variations, we used the databases of dbSNP (24) and JSNP (25). Representative SNPs were mapped onto genes and proteins based on the SNP loci and identifier (rs numbers). Total number of representative SNPs was over 14.5-million. The number of SNPs in the genic region was 5 766 017, where 91 038 SNPs were non-synonymous. Among the amino acid changes caused by nsSNPs, changes in glycine affect protein structure and function most dramatically. Glycines at certain position are strongly conserved evolutionarily due to the size restriction in protein structure. Mutations at such sites would affect the structure and function of the protein significantly (26). We examined the mutation spectrum of amino acids changes caused by nsSNPs (see the website for detailed result), and found a total of 5034 (6.2%) glycine changes due to nsSNPs. In an effort to predict the functional aspects of these nsSNPs, we have analyzed the disease risk for 91 038 nsSNPs using polymorphism phenotyping (PolyPhen) (27).
Modeling structural changes in protein due to genetic variations
To predict structural changes in the drug-related proteins, we have selected 2486 proteins out of 10 883 disease-related proteins that showed sequence similarity over 95% identity with the drug target sequences in the DrugBank database. Search for structural templates for homology modeling was carried out using the BLASTP and PSI-BLAST methods with the minimum percent identity of 60% for the proteins in the PDB structure database (28). We filtered out templates with less than 100 amino acids. This procedure produced the structural templates for 601 drug-related proteins.
Among the candidate templates that covered the nsSNP positions, we selected the template with the highest identity as the primary template. Then the secondary-structure alignment, which is the input for Modeller, was carried out using the local PSI-Pred. Next, we performed 3D structural modeling for drug-related proteins using Modeller (version 9v7) with a single template. Modeller automatically constructs an all-atom 3D model using one or more alignments between the query sequence and the homologous protein sequences of known structure (29). Finally, we determined the best 3D structural model based on the highest stability energy score (z-score).
To examine the structural changes due to amino acid substitution, we generated 4020 mutant proteins at known nsSNP sites. Structural modeling for mutant proteins was carried out in a similar fashion using the same template as the wild-type proteins (see Supplementary Figure S1 for more details). In summary, we constructed 3D structural models for 590 wild-type proteins and 4437 mutant proteins from 538 proteins considering the disease-related nsSNPs (see Supplementary Table S3).
Analysis of protein structural changes and docking simulation
We have analyzed the difference in structural stability between wild-type and mutant proteins. The ΔΔG score of each mutant versus wild-type proteins was calculated using the I-mutant program (version 2.0). This program calculates the free energy difference to estimate the stability change due to mutations (30). Positive ΔΔG scores indicate an increased stability. Large values for ΔΔG (absolute value >1) may indicate significant structural changes, which could affect the drug binding by changing the pocket size or shape (30,31).
Previous studies have reported that protein functions are highly dependent on physical, chemical and geometric features of pockets on the surface of the protein (32,33). Changes in pocket size or stability due to nsSNPs can affect the interactions between target proteins and ligands. Thus, nsSNPs close to the structural pockets are likely to have deleterious effects to be the cause of disease (34) or differences in drug metabolism. To identify the SNP distribution near the pockets, we analyzed the pockets in protein structure using the LIGSITE, which calculates the pocket size and potential ligand-binding sites by the protein–solvent–protein method (35). We examined the pocket sizes up to 10 000 Å3, allowing overlap of maximum three pockets. Most pockets were found to be in the range between 20 and 4000 Å3. More than 50% of nsSNPs were located inside the first two largest pockets.
We also calculated the distances between nsSNPs sites and the structural pockets. It was found that 767 (17%), 2176 (49%) and 3192 (71%) nsSNPs were located within pockets, 5 Å from pockets and 10 Å from pockets, respectively. Because atoms within ∼5–6 Å are able to interact with each other (36), these SNPs can influence interactions between the target protein and ligands.
In an effort to provide the structural picture of drug binding, we performed the docking simulations between the drug with the target and the mutant proteins. Three public programs—AutoDock (version 4.0), Dock (version 6.0) and Fred (version 2.0), were used with the default options and we obtained 981 docking results.
WEB INTERFACE
The VnD web page supports four types of search for user convenience—protein, gene, SNP identifier and disease. Example outputs from the VnD are shown in Figure 2. In the protein menu, users can input a protein ID (UniProt or PDB) and obtain its structural properties, changes by nsSNP(s) and ligand docking information from three public programs. When the number of pockets is clicked, users can observe information about the pockets located in the target protein. Clicking the ‘structure view’ link allows users to observe the protein structure with the Jmol visualization software (http://jmol.sourceforge.net) and download its 3D structural information.
Figure 2.
Query table results and graphic viewer. (a) The server displays information on structures of wild-type and mutant proteins and drug docking found as results for a protein query. The ‘distance between pocket and SNP’ column indicates a pocket located close to a SNP and the distance between a pocket and SNP. The ‘structure view’ column provides the structures of the wild-type and SNP mutant proteins and docking with ligands. (b) Clicking on the gene symbol in the results table (a) allows the user to see the SNP distribution located in the gene structure, protein and disease information.
In the Gene menu, users are able to view the SNP distribution and location in the query gene, related protein information and the relevant disease information as shown in Figure 2b. By clicking the ‘No. of SNPs’ in the ‘Gene Information’ table, information on transcripts and SNP markers is displayed in the GMOD genome browser (37). This would facilitate the recognition of disease-related genetic features such as SNPs within the promoter region or near the splice sites (38).
In the SNP menu, users can obtain detailed information on the SNP including the disease risk estimated from PolyPhen. One can also explore the structural changes in related proteins if the query SNP is nonsynonymous. In addition, the VnD web interface provides a tree view of the disease terms in the UMLS concepts. Currently, the tree view of disease terms consists of 23 top disease terms having an average of five or six sub classes.
To demonstrate the usefulness of the VnD server, we provide the β-2 adrenergic receptor protein (P07550) as an example case. The output pages in Figure 2 can be classified in three categories: (i) physical properties and conformational changes due to nsSNPs in the query protein; (ii) query protein and drug target protein information and (iii) drug ligands and side effect information. Specifically, this query protein is associated with obesity, diabetes, parasitic infection and asthma. The 3D structure and the number of functional sites in the protein are also available in the output. Furthermore, changes in chemical and physical properties such as energy stability caused by six disease-related nsSNPs are also shown. Remarkably, one of the nsSNPs (rs56100672) causes an amino acid substitution (G257R) that changes a small, hydrophobic residue glycine into a polar, bulky, and positively charged residue. The 3D structural models for wild-type and mutant proteins are shown in Supplementary Figure S2. It shows that the pocket size is reduced significantly from 214 to 170 Å3. This size reduction and changes in the pocket shape may have some relationship with the disease and drug susceptibility which need further studies. Therefore, users can observe how the disruption of the surface pocket may affect the protein function and explore its relationship with the molecular causes of a disease or different drug susceptibilities among individuals.
The VnD database server is composed of a web interface and a MySQL (version 5.0.45) database management system. The web interface is implemented in static HTML pages, JSP and Java (version 1.6.0_20). MySQL is used to store the disease-related and drug information.
CONCLUSION
We have constructed a comprehensive database that provides information on genetic variations of disease-related genes and their structural and functional consequences in the aspect of drug target proteins. The effects of non-synonymous SNPs in disease- and drug-related genes were of special focus. We carried out diverse analyses for wild-type and mutant proteins, which include homology modeling, docking, disease risk assessment and analysis on pockets and structural features. Results from all these analyses were integrated into a user-friendly website that would facilitate a mechanistic understanding of trilateral relationships among the genetic variations, diseases and drugs.
The number of disease- and drug-related genes is rapidly increasing partly due to the recent advances in the genome-wide association studies (GWAS). The list of disease-related mutations is expanding as well, as the next-generation sequencing (NGS) techniques become a routine practice. The VnD database will continue to serve as the platform site to explore the relationship between genetic variations and drug effects based on structural modeling and docking simulation.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Korea Research Institute of Bioscience and Biotechnology (KRIBB) Research Initiative Program and ‘Systems Biology Infrastructure Establishment Grant’ provided by Gwangju Institute of Science & Technology in 2010 through Ewha Research Center for Systems Biology (ERCSB). Funding for open access charge: KRIBB Research Initiative Program.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
Authors thank Ms.Eujin Kwak for editing the web figures.
REFERENCES
- 1.The International HapMap Consortium. The International HapMap Project. Nature. 2003;426:789–796. doi: 10.1038/nature02168. [DOI] [PubMed] [Google Scholar]
- 2.Cargill M, Altshuler D, Ireland J, Sklar P, Ardlie K, Patil N, Shaw N, Lane CR, Lim EP, Kalyanaraman N, et al. Characterization of single-nucleotide polymorphisms in coding regions of human genes. Nature Genet. 1999;22:231–238. doi: 10.1038/10290. [DOI] [PubMed] [Google Scholar]
- 3.Yang JO, Hwang S, Oh J, Bhak J, Sohn TK. An integrated database-pipeline system for studying single nucleotide polymorphisms and diseases. BMC Bioinformatics. 2008;9(Suppl. 12):S19. doi: 10.1186/1471-2105-9-S12-S19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Venter JC, Adams MD, Myers EW, Li PW, Mural RJ, Sutton GG, Smith HO, Yandell M, Evans CA, Holt RA, et al. The sequence of the human genome. Science. 2001;291:1304–1351. doi: 10.1126/science.1058040. [DOI] [PubMed] [Google Scholar]
- 5.Chen YH, Liu CK, Chang SC, Lin YJ, Tsai MF, Chen YT, Yao A. GenoWatch: a disease gene mining browser for association study. Nucleic Acids Res. 2008;36:W336–W340. doi: 10.1093/nar/gkn214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Yang IS, Ryu C, Cho KJ, Kim JK, Ong SH, Mitchell WP, Kim BS, Oh HB, Kim KH. IDBD: infectious disease biomarker database. Nucleic Acids Res. 2008;36:D455–D460. doi: 10.1093/nar/gkm925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Goede A, Dunkel M, Mester N, Frommel C, Preissner R. SuperDrug: a conformational drug database. Bioinformatics. 2005;21:1751–1753. doi: 10.1093/bioinformatics/bti295. [DOI] [PubMed] [Google Scholar]
- 9.Voigt JH, Bienfait B, Wang S, Nicklaus MC. Comparison of the NCI open database with seven large chemical structural databases. J. Chem. Inf. Comput. Sci. 2001;41:702–712. doi: 10.1021/ci000150t. [DOI] [PubMed] [Google Scholar]
- 10.Sachs H. Quality control by the Society of hair testing. Forensic Sci. Int. 1997;84:145–150. doi: 10.1016/s0379-0738(96)02057-9. [DOI] [PubMed] [Google Scholar]
- 11.Sheridan RP, Shpungin J. Calculating similarities between biological activities in the MDL Drug Data Report database. J. Chem. Inf. Comput. Sci. 2004;44:727–740. doi: 10.1021/ci034245h. [DOI] [PubMed] [Google Scholar]
- 12.Huey R, Morris GM, Olson AJ, Goodsell DS. A semiempirical free energy force field with charge-based desolvation. J. Comput. Chem. 2007;28:1145–1152. doi: 10.1002/jcc.20634. [DOI] [PubMed] [Google Scholar]
- 13.Bikadi Z, Hazai E. Application of the PM6 semi-empirical method to modeling proteins enhances docking accuracy of AutoDock. J. Cheminformatics. 2009;1:15. doi: 10.1186/1758-2946-1-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.McGaughey GB, Sheridan RP, Bayly CI, Culberson JC, Kreatsoulas C, Lindsley S, Maiorov V, Truchon JF, Cornell WD. Comparison of topological, shape, and docking methods in virtual screening. J. Chem. Inform. Model. 2007;47:1504–1519. doi: 10.1021/ci700052x. [DOI] [PubMed] [Google Scholar]
- 15.Hamosh A, Scott AF, Amberger JS, Bocchini CA, McKusick VA. Online Mendelian Inheritance in Man (OMIM), a knowledgebase of human genes and genetic disorders. Nucleic Acids Res. 2005;33:D514–D517. doi: 10.1093/nar/gki033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Becker KG, Barnes KC, Bright TJ, Wang SA. The genetic association database. Nature Genet. 2004;36:431–432. doi: 10.1038/ng0504-431. [DOI] [PubMed] [Google Scholar]
- 17.Bodenreider O. The Unified Medical Language System (UMLS): integrating biomedical terminology. Nucleic Acids Res. 2004;32:D267–D270. doi: 10.1093/nar/gkh061. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bae JS, Cheong HS, Kim JO, Lee SO, Kim EM, Lee HW, Kim S, Kim JW, Cui T, Inoue I, et al. Identification of SNP markers for common CNV regions and association analysis of risk of subarachnoid aneurysmal hemorrhage in Japanese population. Biochem. Biophys. Res. Commun. 2008;373:593–596. doi: 10.1016/j.bbrc.2008.06.083. [DOI] [PubMed] [Google Scholar]
- 19.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, Dicuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2008;36:D13–D21. doi: 10.1093/nar/gkm1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wheeler DL, Barrett T, Benson DA, Bryant SH, Canese K, Church DM, DiCuccio M, Edgar R, Federhen S, Helmberg W, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2005;33:D39–D45. doi: 10.1093/nar/gki062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kuhn RM, Karolchik D, Zweig AS, Trumbower H, Thomas DJ, Thakkapallayil A, Sugnet CW, Stanke M, Smith KE, Siepel A, et al. The UCSC genome browser database: update 2007. Nucleic Acids Res. 2007;35:D668–D673. doi: 10.1093/nar/gkl928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. UniProtKB/Swiss-Prot: the manually annotated section of the UniProt KnowledgeBase. Methods Mol. Biol. 2007;406:89–112. [Google Scholar]
- 23.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 24.Smigielski EM, Sirotkin K, Ward M, Sherry ST. dbSNP: a database of single nucleotide polymorphisms. Nucleic Acids Res. 2000;28:352–355. doi: 10.1093/nar/28.1.352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hirakawa M, Tanaka T, Hashimoto Y, Kuroda M, Takagi T, Nakamura Y. JSNP: a database of common gene variations in the Japanese population. Nucleic Acids Res. 2002;30:158–162. doi: 10.1093/nar/30.1.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Parrini C, Taddei N, Ramazzotti M, Degl'Innocenti D, Ramponi G, Dobson CM, Chiti F. Glycine residues appear to be evolutionarily conserved for their ability to inhibit aggregation. Structure. 2005;13:1143–1151. doi: 10.1016/j.str.2005.04.022. [DOI] [PubMed] [Google Scholar]
- 27.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, Kondrashov AS, Sunyaev SR. A method and server for predicting damaging missense mutations. Nature Methods. 2010;7:248–249. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berman H, Henrick K, Nakamura H. Announcing the worldwide Protein Data Bank. Nat. Struct. Biol. 2003;10:980. doi: 10.1038/nsb1203-980. [DOI] [PubMed] [Google Scholar]
- 29.John B, Sali A. Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res. 2003;31:3982–3992. doi: 10.1093/nar/gkg460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Capriotti E, Fariselli P, Casadio R. I-Mutant2.0: predicting stability changes upon mutation from the protein sequence or structure. Nucleic Acids Res. 2005;33:W306–W310. doi: 10.1093/nar/gki375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Carey PR, Dong J. Following ligand binding and ligand reactions in proteins via Raman crystallography. Biochemistry. 2004;43:8885–8893. doi: 10.1021/bi049138a. [DOI] [PubMed] [Google Scholar]
- 32.Liu ZP, Wu LY, Wang Y, Chen L, Zhang XS. Predicting gene ontology functions from protein's regional surface structures. BMC Bioinformatics. 2007;8:475. doi: 10.1186/1471-2105-8-475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Laskowski RA, Luscombe NM, Swindells MB, Thornton JM. Protein clefts in molecular recognition and function. Protein Sci. 1996;5:2438–2452. doi: 10.1002/pro.5560051206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Stitziel NO, Binkowski TA, Tseng YY, Kasif S, Liang J. topoSNP: a topographic database of non-synonymous single nucleotide polymorphisms with and without known disease association. Nucleic Acids Res. 2004;32:D520–D522. doi: 10.1093/nar/gkh104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Hendlich M, Rippmann F, Barnickel G. LIGSITE: automatic and efficient detection of potential small molecule-binding sites in proteins. J. Mol. Graph Model. 1997;15:359–363. doi: 10.1016/s1093-3263(98)00002-3. 389. [DOI] [PubMed] [Google Scholar]
- 36.Stitziel NO, Tseng YY, Pervouchine D, Goddeau D, Kasif S, Liang J. Structural location of disease-associated single-nucleotide polymorphisms. J. Mol. Biol. 2003;327:1021–1030. doi: 10.1016/s0022-2836(03)00240-7. [DOI] [PubMed] [Google Scholar]
- 37.Stein LD, Mungall C, Shu S, Caudy M, Mangone M, Day A, Nickerson E, Stajich JE, Harris TW, Arva A, et al. The generic genome browser: a building block for a model organism system database. Genome Res. 2002;12:1599–1610. doi: 10.1101/gr.403602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Abu A, Frydman M, Marek D, Pras E, Stolovitch C, Aviram-Goldring A, Rienstein S, Reznik-Wolf H, Pras E. Mapping of a gene causing brittle cornea syndrome in Tunisian jews to 16q24. Invest. Ophthalmol. Vis. Sci. 2006;47:5283–5287. doi: 10.1167/iovs.06-0206. [DOI] [PubMed] [Google Scholar]