Abstract
The database of three-dimensional interacting domains (3did) is a collection of protein interactions for which high-resolution three-dimensional structures are known. 3did exploits the availability of structural data to provide molecular details on interactions between two globular domains as well as novel domain–peptide interactions, derived using a recently published method from our lab. The interface residues are presented for each interaction type individually, plus global domain interfaces at which one or more partners (domains or peptides) bind. The 3did web server at http://3did.irbbarcelona.org visualizes these interfaces along with atomic details of individual interactions using Jmol. The complete contents are also available for download.
INTRODUCTION
Proteins are key players in virtually all biological events that take place within and between cells. Yet proteins seldom act in isolation and often accomplish their function as part of large molecular machines, whose action is coordinated through intricate regulatory networks of transient protein–protein interactions. It is thus the connections between molecules, rather than the individual components, that will ultimately determine the behaviour of a biological system. Consequently, large resources have been devoted to unveiling protein interrelationships in a high-throughput manner, and the last years have seen the consecution of the first interactome drafts for several model organisms, including human (1–3). However, high-throughput interaction discovery experiments can only indicate that two proteins interact, but do not reveal the molecular details or the mechanism of binding. Currently, this atomic level of detail is only captured in high-resolution three-dimensional (3D) structures, in which individual residue contacts are resolved and the protein interaction interfaces characterized. Efforts to gather and classify such molecular details of interacting structures initially focused on domain–domain interfaces (4,5), while in recent years databases containing structures of more elusive peptide-mediated interactions have been created (6–10).
The database of 3D interacting domains (3did) provides a collection of domain-based interactions for which a high-resolution 3D structure is available. We analysed all interacting structures in the Protein Data Bank (PDB) (11) involving one or more globular domains, and classified them into two main categories on the basis of their contact interfaces: domain–domain and domain–peptide interactions (12). It is worth noting that both classes of interactions can mediate binding between different proteins, but also form intramolecular interactions. Based on the observation that homologous pairs of binding proteins tend to use the same interaction topology (13), we have classified the interactions in 3did by their interface residues. This allows us to provide topological information for each ‘Interaction Type’ (14) as well as global interface regions for each domain by considering all binding partners simultaneously. Where available, we also provide GO terms for the globular domains in 3did (15).
DOMAIN–DOMAIN INTERACTIONS
Globular domains can fold and interact independently from other elements in the same protein and thus constitute ideal modules to drive functional associations of proteins, such as those between different cyclins and CDKs during cell cycle progression (16). Domain–domain interactions form a relatively large and stable interface of ∼2.000 Å2 on average (17). We identified all cases of domain–domain interactions of known 3D structure by first assigning Pfam (18) domains to each individual protein in the PDB using the HMM profiles. Next we computed atomic contacts between domains in the same structure, requiring at least five contacts (hydrogen bonds, electrostatic or van der Waals interactions) to avoid artifacts (non-functional associations) from crystal packing, and removed those lacking a significant interface as described in (19,20). At the moment, there are 159 557 3D structures of domain–domain interactions (DDIs) in 3did, involving 161 996 proteins. These DDIs cover 4186 distinct domains, around a third of the total number of domains in Pfam, forming 5971 different domain pairs between them. The vast majority of these, 4218 DDIs, are always found to mediate binding between different proteins (intermolecular interactions), while 827 are only observed in intramolecular interactions, and 926 additional pairs are found both inter- and intramolecular. It is interesting to note that, in the last two years, the number of unique domain–domain interactions has increased by 20%, thus considerably augmenting the structural coverage of the interaction space.
PEPTIDE-MEDIATED INTERACTIONS
Domain-peptide, or peptide-mediated, interactions occur when a globular domain in one protein recognizes a short linear peptide from another, creating a relatively small interface of ∼350 Å2 on average [according to Stein and Aloy (21)]. This kind of interaction is frequently found in signal transduction networks and sometimes requires dynamic switches like phosphorylation or other post-translational modifications for binding to their recognition domain (22,23). Due to their transient nature, peptide-mediated interactions are more difficult to handle biochemically and thus under-represented in structural databases. The linear motifs that characterize the binding peptide are short patterns of around 10 residues with a common function (i.e. binding to a globular domain) that occur in otherwise unrelated proteins. Despite their shortness, the motifs alone bind their target proteins with sufficient strength to establish a functional interaction (24), while the flanking residues are crucial for specificity (21). Linear motifs are frequently found in disordered or unstructured regions and adopt a well-defined structure only upon binding. In fact, we have exploited this feature to discover ‘hidden’ peptide-mediated interactions among all known 3D structures (see below). A well-studied example of a peptide-mediated interactions occurs between the Src-homology-3 (SH3) domain and proline-rich peptides; [RKY]xxPxxP or PxxPx[KR] are two typical patterns recognized by SH3 domains, where x indicates arbitrary residues and square brackets allow any of the enclosed residues. Much of what is currently known about peptide-mediated interactions is compiled in the Eukaryotic Linear Motif (ELM) database (25), which provides a literature-curated collection of motifs and their interaction partners. In 2008, we published a set of 829 manually curated peptide-mediated interactions in 3D structures matching the patterns in ELM (21), and included these interactions in 3did (7). During the manual curation of these interacting structures, we observed that peptides bound to their recognition domain tend to be more flat and elongated than other peptides of the same length (Figure 1). Based on this characteristic, we created a method to automatically identify peptide-mediated interactions in high-resolution 3D structures, which successfully recognizes known cases as well as novel peptide-binding domains (26). In brief, the method first identifies candidate peptide-domain interactions based on structural features, then these are clustered by interaction topology, and patterns are derived for all clusters with sufficient (non-redundant) information. As an additional validation, we tested whether the derived peptides and their binding domains are significantly over-represented in the current interactomes of human, fly, worm or yeast (26). Only those network-over-represented validated cases are now included in 3did. Due to the automated nature of this method, it will be possible to perform regular updates of the collection of peptide-mediated interactions in 3D structures. Currently, 3did contains 2345 instances of peptide-mediated interactions, involving 1748 protein pairs, 63 Pfam domains and 114 linear motifs either stemming from ELM or derived using the detection method outlined above. This represents roughly a 3-fold increase in the number of peptide-mediated interactions with respect to previous versions of 3did.
Figure 1.
Flat and elongated nature of bound peptides. The chromatin assembly factor 1 peptide [magenta, PDB:1s4z (32)], bound to a chromo shadow domain in HP1beta, illustrates the typical flat and elongated structure that many peptides assume upon binding to their recognition domain. We have recently exploited this feature, together with other structural properties of peptide-mediated interactions, to identify ‘hidden’ cases of such binding events in the PDB (26).
IDENTIFICATION OF INTERACTION INTERFACES
Studying the structures of homologous pairs of interacting proteins has revealed that they very often have the same binding topology (14), although there are exceptions (27). Based on this observation, we have identified and grouped the residues involved in binding interfaces for each domain. In order to get a reference that is stable across the addition of new instances, we aligned all sequences to the HMM profile of the domain, and derived the residue profiles involved in the interactions. Then we clustered the interfaces by the fraction of shared residues as described before (7,26); similar clustering procedures have been implemented for other datasets of interacting protein structures (28,29). As these interaction interfaces are computed for each domain, a domain–domain ‘Interaction Topology’ is classified by the combination of the two interfaces involved. For example, the most common topology for the interaction between Ras and RhoGEF is 0:2, i.e. interface 0 for Ras and interface 2 for RhoGEF, where 0 and 2 are simply identifiers from the clustering procedure (Figure 2). For domain–peptide interactions, only one topology identifier is provided, as we currently do not describe the peptide side of the interface. It should also be noted that not all contacting residues necessarily lie in the HMM profile. In fact, occasionally, none of them does, and in those (rare) cases no interface residues are captured by our method. This implies that the interface positions are not conserved. Interestingly, we find only such interfaces currently in domain–peptide interactions, indicating a lower conservation of these binding sites. According to our current data, the notion still holds that the majority of interaction types always show the same topology (Figure 3). However, for cases with multiple functional interaction topologies it is important to consider these possibilities in applications like homology modelling.
Figure 2.
Domain–domain interactions with interface topologies and structure display. For the selected domain pair, 3did displays all topologies observed in the different instances of this interaction type in all 3D structures, sorted by their frequency. Each ‘Interface Topology’ has an identifier (ID) of the form X:Y that is composed of the respective cluster IDs of the two domains. Here, the most common topology for Ras:RhoGEF is 0:2, while all other observed topologies are much less frequent. For homomeric interactions, X:X marks a symmetric topology. The ‘rainbow’ color scheme is used to indicate where interface residues lie in the sequence, from N-terminus (blue) to C-terminus (red), based on alignment to the HMM profile of the respective domains (see main text). The 3D structure of the selected instance is displayed using Jmol next to the topologies, while the interaction details (PDB ID, domain positions, score, Z-score, topology ID) are listed below. Users can select the 3D structure to be shown by clicking on the Jmol icon in the corresponding row. In domain–domain interactions, the two domains are shown in magenta and cyan, while peptide-binding domains are shown in a rainbow colour scheme to match the interface visualization.
Figure 3.
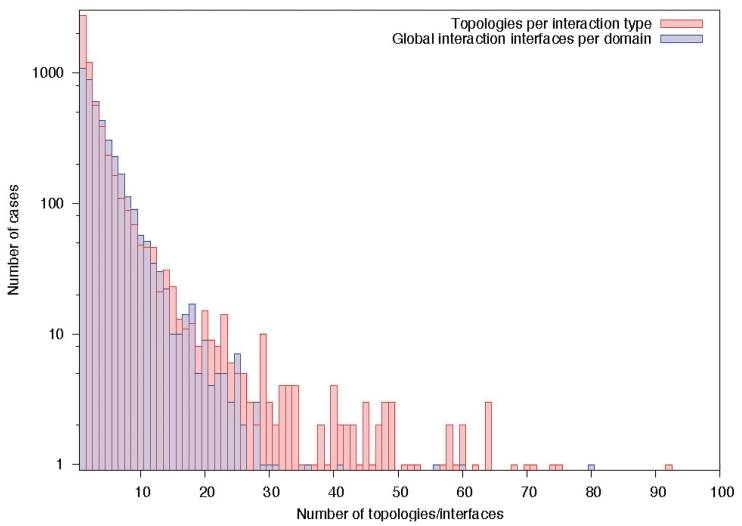
Number of domain interfaces. About half of the domain–domain interactions currently stored in 3did are only observed in one interaction topology, and only a small fraction shows ten or more different binding orientations. Similarly, roughly 50% of the domains in 3did have one or two global binding interfaces, while few have 10 interfaces or more.
In addition to the individual interfaces for each interaction type, we have now introduced global interface clusters for each domain. These group binding partners use the same, or largely overlapping, interaction surfaces of a given domain, and may thus help identify positions which are crucial for binding multiple partners. This is especially important for proteins like Ras, which have many binding partners with overlapping interfaces (13,30). The global interfaces are computed via complete linkage hierarchical clustering (31) over the fractions of overlapping positions in all individual interaction-type-interfaces for this domain. Cases with a minimum overlap of 25% among all partners are grouped together. In the interface visualization of the 3did web server, the fraction of different partners using a given residue is indicated by the height of the corresponding bar. At the moment, we find multiple binding partners for 4020 interaction interfaces on 1675 domains in 3did. Overall, 2511 domains have only single-partner interfaces, 162 have only multi-partner interfaces and 1513 have both types (Figure 3).
3did USAGE AND VISUALIZATION
The easiest way of searching 3did for domain-based interactions is through our web interface, which can be queried with a domain or motif name or keyword, a pair of interacting or indirectly connected domains, the sequence or PDB code of a particular protein, or GO terms of interest. As in previous versions, 3did will then display all domains and peptides that physically interact with the domain of interest (or present in the selected structure or sequence) and for which 3D data of the interaction interface is available. Interaction partners are grouped by the global binding interfaces described above, which are visualized in a blue-to-red rainbow colour scheme (from N- to C-terminus). The relative height of the individual position bars indicates the fraction of binding partners that actually use this residue (Figure 4). If only one binding partner is found for a given interface, all these bars have the same height. Note that, as these interfaces are based on Pfam HMMs, a sequence of interest needs to be aligned to the respective HMM in order to identify the interacting residues. All interaction partners will also be displayed in an interactive network indicating the type of element (domain or peptide), whether the interactions are intra or intermolecular, and functional annotations from GO where available. From the list of interacting domains and peptides, the user can select individual interaction types to access their molecular details. The domain–domain and domain–peptide interaction pages displays all interface topologies of the domain(s) involved in the binding, along with the frequency of their occurrence in the current set of 3D structures. As described above, for domain–domain interactions the ‘Interaction Topology’ is composed of the two interface IDs involved (cf. Figure 2). The interaction pages also provide listings of each 3D structure in which the selected interaction type is found, plus detailed information on the position of the domains and peptides in this structure. Furthermore, it provides empirical potential scores and Z-scores for the interaction, which indicates the number of favourable contacting residue pairs in this interface (19,20). In general, the higher the Z-score the more specific an interaction is. The actual 3D structure of the interaction is displayed in the upper right corner of the page by clicking on the Jmol (http://www.jmol.org) icon (Figure 2). For domain–domain interactions, the two domains are coloured magenta and cyan and shown in ‘cartoon’ representation with the residues participating in the interface (i.e. making hydrogen bonds, salt bridges or van der Waals contacts) shown as ‘sticks’. For domain–peptide interactions, the domain is in ‘cartoon’ representation and coloured following a ‘rainbow’ scheme that corresponds to the HMM-profile-based visualization of the interface residues, while the peptide is shown in gray, and interacting residues are again shown as ‘sticks’. For entire PDB structures, all chains are shown in ‘cartoon’ representation.
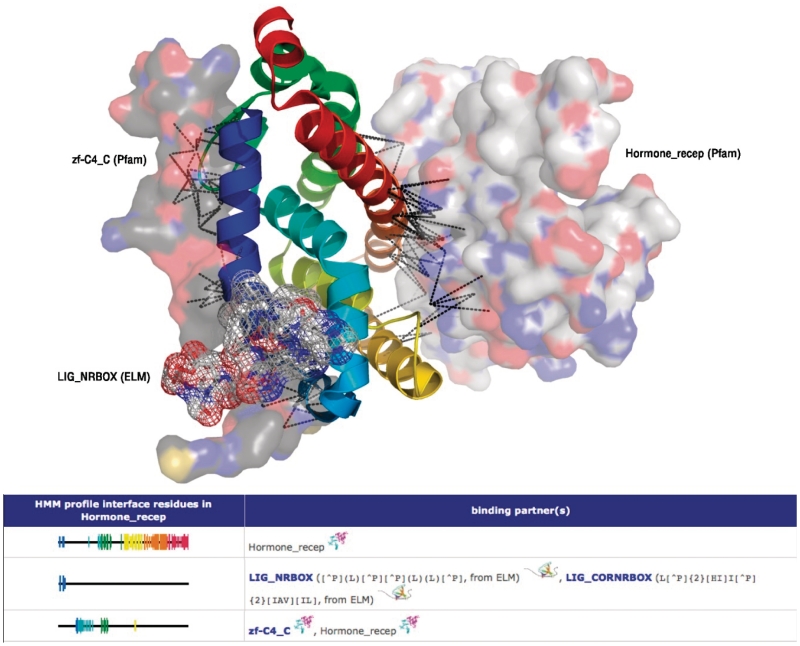
Figure 4.
Multiple binding interfaces and their visualization. The upper part of the figure shows the hormone_recep domain (‘cartoon’ representation, rainbow colour scheme) and three of its binding partners, two domains (‘surface’ representation) and one linear motif (‘mesh’ representation). Below, the global interfaces are visualized as on the web page. These interfaces group binding partners using the same or largely overlapping interfaces, and indicates which profile positions are involved in the binding. This allows quick identification of possible overlaps, and thus competition, of different binding partners. Like the individual topologies, global interfaces are displayed in a blue-to-red rainbow colour scheme. The height of the position bars indicates how many binding partners use this particular position. By clicking on a binding partner, the user is redirected to that element’s page, while the interaction symbol (two domains or a domain and a peptide) leads to the respective interaction page (cf. Figure 2).
AVAILABILITY
The 3did web server at http://3did.irbbarcelona.org allows direct querying of the database and provides MySQL dumps and flat files containing the full dataset for download, for users interested in large-scale studies. Domain–domain interactions in 3did are updated weekly to include newly released structures. Peptide-mediated interactions will be updated in major releases, which will occur when new Pfam versions become available.
FUNDING
Grants BIO2007-62426 and PSS-010000-2009 from the Spanish Ministerio de Educación y Ciencia and the European Commission under FP7 Grant Agreement 223101(AntiPathoGN). Funding for open access charge: BIO2007-62426.
Conflict of interest statement. None declared.
REFERENCES
- 1.Rual JF, Venkatesan K, Hao T, Hirozane-Kishikawa T, Dricot A, Li N, Berriz GF, Gibbons FD, Dreze M, Ayivi-Guedehoussou N, et al. Towards a proteome-scale map of the human protein–protein interaction network. Nature. 2005;437:1173–1178. doi: 10.1038/nature04209. [DOI] [PubMed] [Google Scholar]
- 2.Stelzl U, Worm U, Lalowski M, Haenig C, Brembeck FH, Goehler H, Stroedicke M, Zenkner M, Schoenherr A, Koeppen S, et al. A human protein–protein interaction network: a resource for annotating the proteome. Cell. 2005;122:957–968. doi: 10.1016/j.cell.2005.08.029. [DOI] [PubMed] [Google Scholar]
- 3.Ewing RM, Chu P, Elisma F, Li H, Taylor P, Climie S, McBroom-Cerajewski L, Robinson MD, O'Connor L, Li M, et al. Large-scale mapping of human protein–protein interactions by mass spectrometry. Mol. Syst. Biol. 2007;3:89. doi: 10.1038/msb4100134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Stein A, Russell RB, Aloy P. 3did: interacting protein domains of known three-dimensional structure. Nucleic Acids Res. 2005;33:D413–D417. doi: 10.1093/nar/gki037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Winter C, Henschel A, Kim WK, Schroeder M. SCOPPI: a structural classification of protein–protein interfaces. Nucleic Acids Res. 2006;34:D310–D314. doi: 10.1093/nar/gkj099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ceol A, Chatr-aryamontri A, Santonico E, Sacco R, Castagnoli L, Cesareni G. DOMINO: a database of domain–peptide interactions. Nucleic Acids Res. 2007;35:D557–D560. doi: 10.1093/nar/gkl961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Stein A, Panjkovich A, Aloy P. 3did Update: domain–domain and peptide–mediated interactions of known 3D structure. Nucleic Acids Res. 2009;37:D300–D304. doi: 10.1093/nar/gkn690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Encinar JA, Fernandez-Ballester G, Sánchez IE, Hurtado-Gomez E, Stricher F, Beltrao P, Serrano L. ADAN: a database for prediction of protein–protein interaction of modular domains mediated by linear motifs. Bioinformatics. 2009;25:2418–2424. doi: 10.1093/bioinformatics/btp424. [DOI] [PubMed] [Google Scholar]
- 9.Vanhee P, Reumers J, Stricher F, Baeten L, Serrano L, Schymkowitz J, Rousseau F. PepX: a structural database of non-redundant protein–peptide complexes. Nucleic Acids Res. 2010;38:D545–D551. doi: 10.1093/nar/gkp893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.London N, Movshovitz-Attias D, Schueler-Furman O. The structural basis of peptide–protein binding strategies. Structure. 2010;18:188–199. doi: 10.1016/j.str.2009.11.012. [DOI] [PubMed] [Google Scholar]
- 11.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Aloy P, Russell RB. Structural systems biology: modelling protein interactions. Nat. Rev. Mol. Cell Biol. 2006;7:188–197. doi: 10.1038/nrm1859. [DOI] [PubMed] [Google Scholar]
- 13.Aloy P, Ceulemans H, Stark A, Russell RB. The relationship between sequence and interaction divergence in proteins. J. Mol. Biol. 2003;332:989–998. doi: 10.1016/j.jmb.2003.07.006. [DOI] [PubMed] [Google Scholar]
- 14.Aloy P, Russell RB. Ten thousand interactions for the molecular biologist. Nat. Biotechnol. 2004;22:1317–1321. doi: 10.1038/nbt1018. [DOI] [PubMed] [Google Scholar]
- 15.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, et al. Gene ontology: tool for the unification of biology. The Gene Ontology Consortium. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jeffrey PD, Russo AA, Polyak K, Gibbs E, Hurwitz J, Massagué J, Pavletich NP. Mechanism of CDK activation revealed by the structure of a cyclinA-CDK2 complex. Nature. 1995;376:313–320. doi: 10.1038/376313a0. [DOI] [PubMed] [Google Scholar]
- 17.Chakrabarti P, Janin J. Dissecting protein–protein recognition sites. Proteins. 2002;47:334–343. doi: 10.1002/prot.10085. [DOI] [PubMed] [Google Scholar]
- 18.Finn RD, Mistry J, Tate J, Coggill P, Heger A, Pollington JE, Gavin OL, Gunasekaran P, Ceric G, Forslund K, et al. The Pfam protein families database. Nucleic Acids Res. 2010;38:D211–D222. doi: 10.1093/nar/gkp985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aloy P, Russell RB. Interrogating protein interaction networks through structural biology. Proc. Natl Acad. Sci. USA. 2002;99:5896–5901. doi: 10.1073/pnas.092147999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Aloy P, Russell RB. InterPreTS: protein interaction prediction through tertiary structure. Bioinformatics. 2003;19:161–162. doi: 10.1093/bioinformatics/19.1.161. [DOI] [PubMed] [Google Scholar]
- 21.Stein A, Aloy P. Contextual specificity in peptide-mediated protein interactions. PLoS ONE. 2008;3:e2524. doi: 10.1371/journal.pone.0002524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Pawson T. Dynamic control of signaling by modular adaptor proteins. Curr. Opin. Cell Biol. 2007;19:112–116. doi: 10.1016/j.ceb.2007.02.013. [DOI] [PubMed] [Google Scholar]
- 23.Stein A, Pache RA, Bernadó P, Pons M, Aloy P. Dynamic interactions of proteins in complex networks: a more structured view. FEBS J. 2009;276:5390–5405. doi: 10.1111/j.1742-4658.2009.07251.x. [DOI] [PubMed] [Google Scholar]
- 24.Kim HY, Ahn BY, Cho Y. Structural basis for the inactivation of retinoblastoma tumor suppressor by SV40 large T antigen. Embo J. 2001;20:295–304. doi: 10.1093/emboj/20.1.295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gould CM, Diella F, Via A, Puntervoll P, Gemund C, Chabanis-Davidson S, Michael S, Sayadi A, Bryne JC, Chica C, et al. ELM: the status of the 2010 eukaryotic linear motif resource. Nucleic Acids Res. 2010;38:D167–D180. doi: 10.1093/nar/gkp1016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Stein A, Aloy P. Novel peptide-mediated interactions derived from high-resolution 3-dimensional structures. PLoS Comput. Biol. 2010;6:e1000789. doi: 10.1371/journal.pcbi.1000789. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Park SY, Beel BD, Simon MI, Bilwes AM, Crane BR. In different organisms, the mode of interaction between two signaling proteins is not necessarily conserved. Proc. Natl Acad. Sci. USA. 2004;101:11646–11651. doi: 10.1073/pnas.0401038101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim WK, Henschel A, Winter C, Schroeder M. The many faces of protein–protein interactions: a compendium of interface geometry. PLoS Comput. Biol. 2006;2:e124. doi: 10.1371/journal.pcbi.0020124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Teyra J, Paszkowski-Rogacz M, Anders G, Pisabarro MT. SCOWLP classification: structural comparison and analysis of protein binding regions. BMC Bioinformatics. 2008;9:9. doi: 10.1186/1471-2105-9-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kiel C, Beltrao P, Serrano L. Analyzing protein interaction networks using structural information. Annu. Rev. Biochem. 2008;77:415–441. doi: 10.1146/annurev.biochem.77.062706.133317. [DOI] [PubMed] [Google Scholar]
- 31.de Hoon MJ, Imoto S, Nolan J, Miyano S. Open source clustering software. Bioinformatics. 2004;20:1453–1454. doi: 10.1093/bioinformatics/bth078. [DOI] [PubMed] [Google Scholar]
- 32.Thiru A, Nietlispach D, Mott HR, Okuwaki M, Lyon D, Nielsen PR, Hirshberg M, Verreault A, Murzina NV, Laue ED. Structural basis of HP1/PXVXL motif peptide interactions and HP1 localisation to heterochromatin. EMBO J. 2004;23:489–499. doi: 10.1038/sj.emboj.7600088. [DOI] [PMC free article] [PubMed] [Google Scholar]