Abstract
BTO, the BRENDA Tissue Ontology (http://www.BTO.brenda-enzymes.org) represents a comprehensive structured encyclopedia of tissue terms. The project started in 2003 to create a connection between the enzyme data collection of the BRENDA enzyme database and a structured network of source tissues and cell types. Currently, BTO contains more than 4600 different anatomical structures, tissues, cell types and cell lines, classified under generic categories corresponding to the rules and formats of the Gene Ontology Consortium and organized as a directed acyclic graph (DAG). Most of the terms are endowed with comments on their derivation or definitions. The content of the ontology is constantly curated with ∼1000 new terms each year. Four different types of relationships between the terms are implemented. A versatile web interface with several search and navigation functionalities allows convenient online access to the BTO and to the enzymes isolated from the tissues. Important areas of applications of the BTO terms are the detection of enzymes in tissues and the provision of a solid basis for text-mining approaches in this field. It is widely used by lab scientists, curators of genomic and biochemical databases and bioinformaticians. The BTO is freely available at http://www.obofoundry.org.
INTRODUCTION
Ontologies which are used in life science represent classification systems that provide a controlled vocabulary for a biological or biomedical knowledge domain. They are flexibly organized to cope with an increasing amount of information in a structured way. The vocabulary items constitute a single common set of terms that enables the use of a formal unified terminology. The terms are connected among each other through well defined relationships. These ‘parent–child’ relationships permit the depiction of the hierarchical structure of the ontology which contains terms at various levels of detail. An important pioneering effort in the field of biological ontologies, probably being the most widely used, is the Gene Ontology (GO) that aims at a standardized functional description of genes and their products (1).
The BRENDA Tissue Ontology (BTO) (http://www.BTO.brenda-enzymes.org) was initiated in 2003 to develop a standardized representation of all tissue terms from every taxonomic group covering animals, plants, fungi and prokaryotes which are connected to enzyme data in the BRENDA enzyme database (2). The first version was described in brief in a publication on the BRENDA enzyme resource in 2004 (3). The increasing amount of enzyme data and the construction of flexible query options demanded the development of a hierarchical ontology of tissues and cell types representing the sources of enzymes restricted to specific tissues or organs. This vocabulary also includes tissues and organs that are specific to taxonomic groups or single species. Since the development of the Gene Ontology (1,4) as the major collaborative project to standardize the representation and annotation of genes and their products, many biological ontologies have emerged. Most of them are associated with the Open Biological and Biomedical Ontologies Foundry [OBO, (5)] and are freely available from its website (http://www.obofoundry.org). They include anatomical and developmental ontologies that exclusively focus on various model organisms such as mouse, Drosophila melanogaster or Arabidopsis thaliana. The Cytomer database provides an overview on expression sources such as organs, tissue, cell types and developmental stages, focusing on the human system (6). In contrast, the Cell Ontology (7) and the eVOC Ontologies (8) integrate all organisms, but they focus solely on cell types. The Plant Ontology database [PO, (9)] provides a complex hierarchical structure of botanical terms with controlled vocabularies in the annotation of plant-related tissues, growth stage specific expression of genes, proteins and phenotypes. However, it does not support other taxonomic groups such as animals and fungi. Furthermore, the cellular component sub-ontology of GO is restricted to the sub-cellular level and does not extend to multi-cellular structures such as tissues or organs.
In this article, we describe the BTO as an integrating dictionary for enzymes sources, its content and characteristics, the web interface and the usage of this comprehensive structured encyclopedia of organism-specific tissue terms linked to enzyme functional data. The BTO has been developed according to the rules and formats of the GO Consortium and provides the first ontology for all organisms with respect to the diversity of enzyme sources.
STRUCTURE OF THE BTO
BTO terms: the nodes of the graph
All manually extracted enzyme source tissue and organ terms were evaluated and then classified into the hierarchical structure of the ontology. Like the GO, the BTO is organized as a directed acyclic graph (DAG) whose nodes are represented by the BTO terms (Figure 1). The ontology was constructed using the open source Java tool OBO-Edit (formerly known as DAG-Edit), developed by the GO Consortium. Every term (e.g. epithelium) occurs only once in the ontology, hence the entirety of terms is a true set according to the mathematical definition. The terms have definitions and textual descriptions. One or more references lead to the source of information. Each term possesses at least one relationship to another term (see below). As unique identifier each term has a condensed zero-padded seven digit identifier prefixed by ‘BTO:’. These unique identifiers are stored in a relational database (MySQL) and serve as stable accession numbers in order to establish cross references to biochemical databases such as BRENDA.
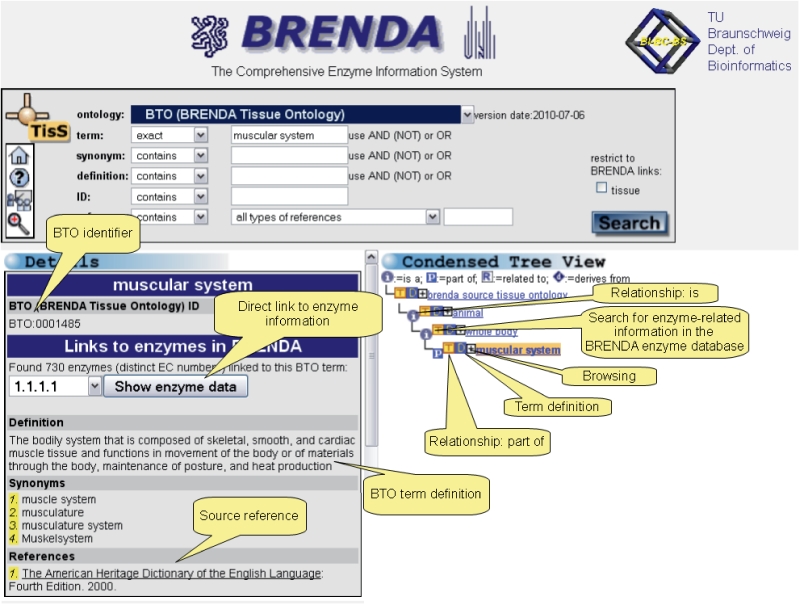
Figure 1.
Web Interface with search and display capabilities of the BTO. As an example, the term ‘muscular system’ was chosen. The ‘condensed tree view’ provides an overview of the position of the term of interest in the hierarchical structure of the BTO.
Relationships between the terms: the edges of the graph
The actual structure of a graph is represented by the relationships between its nodes: the edges. In biological ontologies, the edges describe ‘parent–child’ relationships between the controlled vocabulary terms. For an accurate description of biological ontologies such as the BTO the need for different types of relationships has to be considered in order to correctly dissolve the relationship between the ‘parent’ and ‘child’ terms. Four different types of ‘parent–child’ relationships are defined in the BTO (Figure 2).
is_a
e.g. cardiac muscle fibre is_a muscle fibre
part_of
e.g. muscle fibre is part_of muscle
develops_from/derives_from
e.g. myoma cell develops_from/derives_from muscle fibre
related_to
e.g. electroplax is related_to muscle fibre
The relationship type ‘related_to’ was established to describe more general relationships between tissue terms which cannot be defined using the other ones. An example is given by the relationship ‘electroplax’ and ‘muscle fibre’. The term ‘electroplax’ is defined as: ‘A stack of specialized muscle fibres found in electric eels, arranged in series. The fibres have lost the ability to contract, instead they generate extremely high voltages (ca. 500 V) in response to nervous stimulation. They contain asymmetrically distributed sodium potassium ATPases, acetylcholine receptors and sodium gates at extraordinarily high concentrations’.
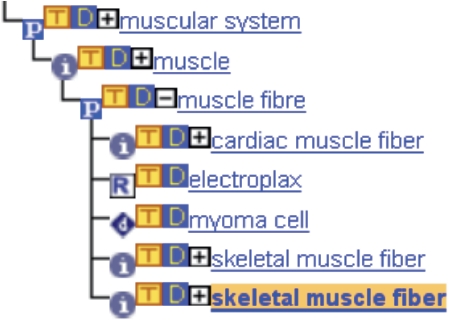
Figure 2.
Ontology relationships for ‘muscle fibre’ and its descendants, whereas the term ‘muscle fibre’ is a ‘part_of’ a ‘muscle’ (Symbol: P) and a ‘myoma cell’ ‘develops_from’ a ‘muscle fibre’ (Symbol: d). In contrast, the parent–child relationship between ‘muscle fibre’ and ‘electroplax’ is very general represented by the relationship type ‘related_to’ (Symbol: R).
CONTENT OF THE BTO AND DATA ANNOTATION
The BTO draws upon the comprehensive enzyme-related data repository of the BRENDA enzyme database, including information on the occurrence of the enzyme source: the anatomical structures, tissues, cell lines, cell types, cancerous tissues from uni- and multi-cellular organisms such as prokaryotes, mammalia, plants, fungi or viruses. Currently, BRENDA contains ∼75 000 enzyme-organism-specific tissue entries updated twice yearly (BRENDA release 2010.2). These entries were manually extracted from more than 100 000 different literature references. Besides that, terms and concepts from external sources such as UniProt (10), the Experimental Factor Ontology [EFO, (11)], the Foundational Model of Anatomy ontology [FMA, (12)] and the PAZAR Project (13) are integrated into the BTO.
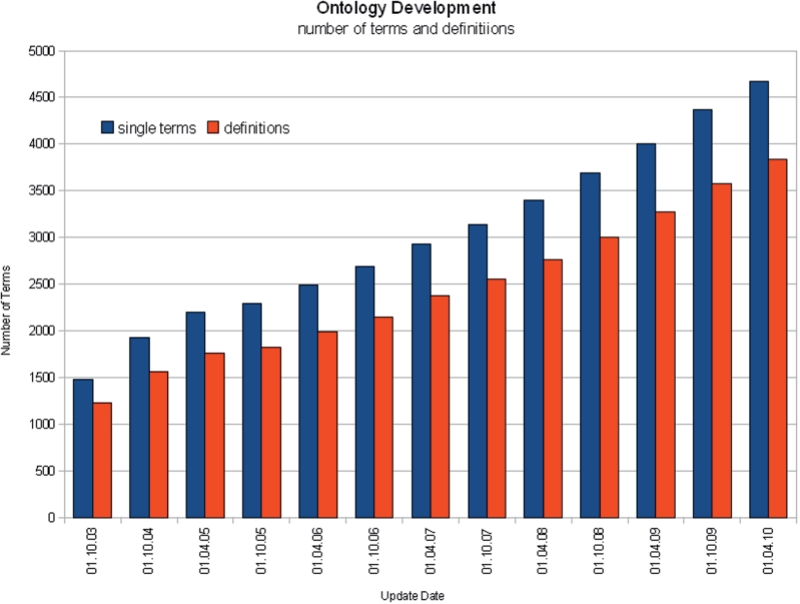
Since 2003, the number of terms in the BTO increased to 4724 (Figure 3) and the number of all entries, including the synonyms, increased to 8287. The ontology is updated biannually. After each update the data increases by 500–600 different terms.
Figure 3.
Number of BTO terms since 2003.
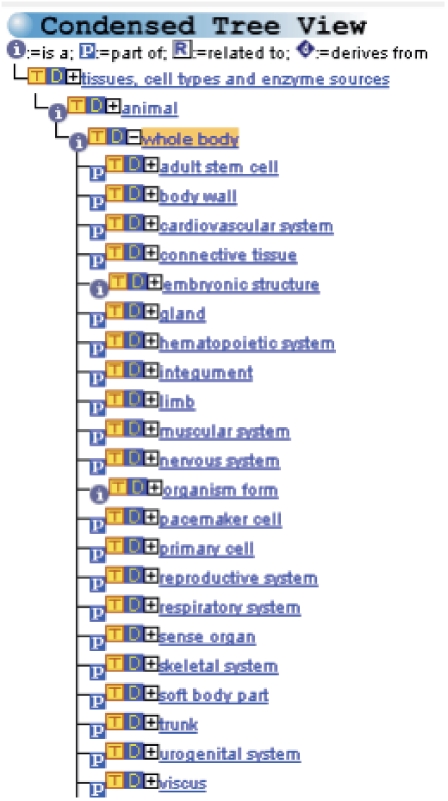
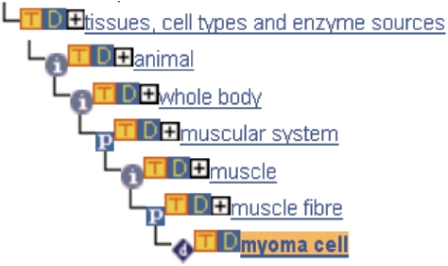
The terms are classified into four main categories, which are represented as four separated, non-overlapping subgraphs: animal, plant, fungus and ‘other sources’. For example, the term ‘whole body’, a child term of ‘animal’ has 22 direct child terms (Figure 4). These terms have in total 4142 descendant terms (child terms, grandchild terms, etc.). Furthermore, terms representing cell types are assigned to the tissues from which they originate or to which they are related. Therefore, the term ‘myoma cell’ (a muscular tumour cell) is assigned to the main category ‘animal’ and to the sublevel ‘muscular system’ for example (Figure 5).
Figure 4.
BTO subgraph for ‘Animal’ with its direct child terms.
Figure 5.
The assignment of ‘myoma cell’ in the BTO.
Most of the terms of different organisms are distinguished by the connection of the tissue or cell type to the associated organism information. However, there may be several identical designations for tissues both in plants and animals, e.g. ‘epidermis’. To distinguish between those tissue terms and to assign them correctly into the ontology for plant tissues the prefix ‘plant’ is inserted in front of the term, e.g. ‘plant epidermis’.
Additionally, the BTO contains disease-related tissue terms. For example, the term ‘Alzheimer specific cell type’ was introduced to classify the abnormally developed brain tissues in Alzheimer’s disease. This term was assigned as a child to the term ‘cerebral cortex’ with the relationship type ‘related to’. Similarly for epithelioma (a specific type of epithelial cancer) the term ‘epithelioma cell’ classified as ‘derived from epithelial cell’ has been embedded. Another example is ‘cystic fibrosis disease specific cell type’ with the parent term ‘exocrine gland’.
Since abbreviations are commonly used in the laboratories and subsequently also adopted in the scientific publications, cell line names often consist of short letter–figure combinations, e.g. ‘A6 cell’, ‘L6 cell’ or ‘A-14 cell’. To avoid inconsistencies and ambiguities those terms are renamed within the BTO and described in more detail by checking the original literature reference. For example, ‘A6 cell’ is replaced by ‘Xenopus A6 cell’, ‘L6 cell’ by ‘L6 myoblast cell’ and ‘A-14 cell’ by ‘3T3-A14 cell’. Other short letter combinations such as ‘OEC’ could have multiple meanings, standing for ‘ovarian epithelial cell’, ‘olfactory ensheathing cell’ and also ‘oral epithelial cell’. Therefore, the respective unabridged wording is chosen as the BTO term and ‘OEC’ is included as synonym for all of them.
Increased annotation efforts in specific emerging fields of research
The recent focus on specific fields of research in the scientific community, i.e. cancer research, brain research or stem cell research is reflected in an increase in the number of terms in the respective branches of the BTO. The major part of recently added BTO terms are newly created cell lines which have been established in many different laboratories. Some of these are also indexed in the large cell line databases such as ATCC—American Cell Type Culture Collection (http://www.atcc.org), ECACC—European Collection of Cell Cultures (http://www.hpacultures.org.uk/collections/ecacc.jsp) or DSMZ (http://www.dsmz.de). In this manner 96 new melanoma cell lines have been annotated in the last year.
Enzymes involved in brain function have also gained increased interest of researchers. This is reflected in a growing number of brain-related terms. Currently the BTO contains 218 distinct brain-related terms. These terms encompass various brain areas and are classified according to their anatomical and functional structures. Many general terms have been supplemented with new specific child terms in this context. For example the term ‘neuron’ meanwhile has 34 child terms, 11 of which are neuronal stem cells. These cell types have all been described as enzyme sources.
Efforts in finding definitions for terms
More than 80% of the tissue terms are associated with a definition that concisely describes the meaning and context of the term and are linked to one or more respective references. Whenever available, internationally accepted definitions obtained from medical dictionaries, cell line databases or other expert dictionaries were entered such as Dorlands Medical Dictionary [http://www.dorlands.com, (14)], NCI Dictionary of Cancer Terms (http://www.cancer.gov/dictionary), ATCC, ECACC or Merriam Websters Dictionary [http://www.merriam-webster.com, (15]. Terms without a definition can be found in two categories:
generic parent terms which do not need a definition, e.g. gastric cancer cell line, as a parent term for various cell lines; and
culture condition terms defining a compound which must be present in the culture medium for the induction of the enzyme, e.g. ‘culture condition: D-xylose grown cell’. D-xylose 1-dehydrogenase is expressed in Arthrobacter or Haloarcula only if D-xylose is added to the growth medium.
WEB INTERFACE AND AVAILABILITY
As part of the BRENDA enzyme database, all entries of the BTO are also stored in a relational database. Several web-based search options are provided to access the entries of the BTO via the BRENDA web site.
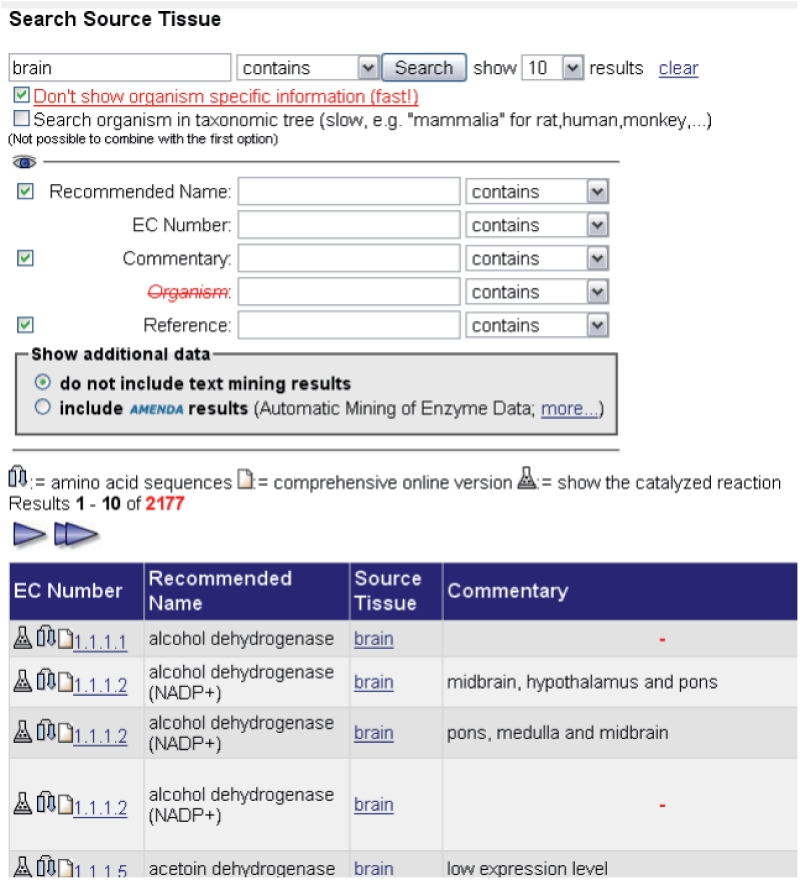
The enzyme sources can be searched via the BRENDA ‘Quick Search’ mode using the Source Tissue search form (see Figure 6, http://www.brenda-enzymes.org/index.php4?page=/php/search_result.php4?a=33) or the ‘Advanced Search’ (http://www.brenda-enzymes.org/index.php4?page=adv_search/index.php4). As a result of a ‘Quick Search’, the user receives a list of all enzymes which are isolated from or detected in the searched BTO tissue. In the next step, the user can directly move on to the BTO website (Figure 1), by clicking on the BTO term or can obtain more detailed information from the comprehensive enzyme result view by clicking on the EC number.
Figure 6.
The ‘Source Tissue’ search form of the BRENDA web interface. As an example, parts of the search results for the tissue term ‘brain’ are shown (enzyme hits).
In addition, there is another versatile web interface (http://www.BTO.brenda-enzymes.org) that offers additional search and navigation functionalities within the BTO (Figure 1). It offers a search for BTO terms, synonyms, definitions or references. A combined search using several of these fields with the boolean operator ‘AND’ is also possible.
As a result, the graphical representation of the searched term in the form of tree-like subgraph of the BTO is displayed. The frame ‘condensed tree view’ provides an overview of the position of the term of interest in the hierarchical structure of the BTO (Figure 1). Here, the predecessor terms up to the root are shown. Furthermore, the user is enabled to display all direct child terms of the selected term, display the definitions of the terms and easily identify the relationship type between two nodes. Moreover, all enzymes that are related to the selected BTO term are displayed in a selection field. These comprise, for example enzymes that are isolated from the respective tissue or organ. The listed EC numbers are directly connected to the enzyme information of the BRENDA database.
It is also possible to search for enzymes isolated from a specific BTO tissue, and—if desired—all of its child and related terms using the symbol  in the graphical presentation. For example, the search for ‘forebrain’ alone and with its ramifications yields 35 and 1239 hits, respectively (Figure 7).
in the graphical presentation. For example, the search for ‘forebrain’ alone and with its ramifications yields 35 and 1239 hits, respectively (Figure 7).
Figure 7.
Search for the term ‘forebrain’ in the BRENDA enzyme database.
Via the web interface, the BTO can be freely downloaded as a text file from the BRENDA web site (http://www.brenda-enzymes.org) or in the OBO and OWL format from http://www.obofoundry.org/cgi-bin/detail.cgi?id=brenda. The file can be visualized with tools such as OBO-Edit and integrated into a database system for own purposes.
USAGE OF THE BTO IN THE COMMUNITY
The BTO is widely used in the scientific community. Queries in web search engines yield ∼10 000 hits for example. Several secondary databases make use of the BTO. The Tissue DistributionDBs (16) uses the controlled vocabulary terms of the BTO to create an organism-specific repository of tissue distribution profiles for identifying and ranking the genes based on Expressed Sequence Tags (ESTs). The PRoteomics IDEntifications database [PRIDE, (17)], the main data repository of proteomics data and also the PAZAR project use the BTO as a reference to define and specify tissues and cell types. The Genes-to-Systems Breast Cancer (G2SBC) database (18), an online resource for molecular and systems biology of breast cancer information also includes the BTO within their project.
FUTURE ENHANCEMENTS
The BTO is currently designed as a human-readable hierarchical vocabulary of enzyme-containing tissues, which is already widely used in biochemical applications. Making it purpose-independent and to include terms that are not connected to enzymes would allow an even larger and wider application and could increase its value for text mining procedures.
FUNDING
This work was supported by the European Union: (FELICS: Free European Life-Science Information and Computational Services: 021902 (RII3); SLING: Serving Life-science Information for the Next Generation: 226073).
Conflict of interest statement. None declared.
REFERENCES
- 1.Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight S, Eppig JT, et al. Gene ontology: tool for the unification of biology. Nat. Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Chang A, Scheer M, Grote A, Schomburg I, Schomburg D. BRENDA, AMENDA and FRENDA the enzyme information system: new content and tools in 2009. Nucleic Acids Res. 2009;37:D588–D592. doi: 10.1093/nar/gkn820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Schomburg I, Chang A, Ebeling C, Gremse M, Heldt C, Huhn G, Schomburg D. BRENDA, the enzyme database: updates and major new developments. Nucleic Acids Res. 2004;32:D431–D433. doi: 10.1093/nar/gkh081. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.The Gene Ontology Consortium. The Gene Ontology in 2010: extensions and refinements. Nucleic Acids Res. 2010;38:D331–D335. doi: 10.1093/nar/gkp1018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Smith B, Ashburner M, Rosse C, Bard C, Bug W, Ceusters W, Goldberg LJ, Eilbeck K, Ireland A, Mungall CJ, et al. The OBO Foundry: coordinated evolution of ontologies to support biomedical data integration. Nat. Biotechnol. 2007;25:1251–1255. doi: 10.1038/nbt1346. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Michael H, Chen X, Fricke E, Haubrock M, Ricanek R, Wingender E. Deriving an ontology for human gene expression sources from the CYTOMER database on human organs and cell types. In Silico. 2005;5:61–66. [PubMed] [Google Scholar]
- 7.Bard J, Rhee SY, Ashburner M. An ontology for cell types. Genome Biol. 2005;6:R21. doi: 10.1186/gb-2005-6-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kelso J, Visagie J, Theiler G, Christoffels A, Bardien-Kruger S, Smedley D, Otgaar D, Greyling G, Jongeneel V, McCarthy M, et al. eVOC: a controlled vocabulary for gene expression data. Genome Res. 2003;13:1222–1230. doi: 10.1101/gr.985203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Avraham S, Tung CW, Ilic K, Jaiswal P, Kellogg EA, McCouch S, Pujar A, Reiser L, Rhee SY, Sachs MM, et al. The Plant Ontology Database: a community resource for plant structure and developmental stages controlled vocabulary and annotations. Nucleic Acids Res. 2008;36:D449–D454. doi: 10.1093/nar/gkm908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.The UniProt Consortium. The Universal Protein Resource (UniProt) in 2010. Nucleic Acids Res. 2010;38:D142–D148. doi: 10.1093/nar/gkp846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Malone J, Holloway E, Adamusiak T, Kapushesky M, Zheng J, Kolesnikov N, Zhukova A, Brazma A, Parkinson H. Modeling sample variables with an Experimental Factor Ontology. Bioinformatics. 2010;26:1112–1118. doi: 10.1093/bioinformatics/btq099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Rosse C, Mejino JVL. A reference ontology for biomedical informatics: the Foundational Model of Anatomy. J. Biomed. Inform. 2003;36:478–500. doi: 10.1016/j.jbi.2003.11.007. [DOI] [PubMed] [Google Scholar]
- 13.Portales-Casamar E, Kirov S, Lim J, Lithwick S, Swanson MI, Ticoll A, Snoddy J, Wasserman WW. PAZAR: a framework for collection and dissemination of cis-regulatory sequence annotation. Genome Biol. 2007;8:R207. doi: 10.1186/gb-2007-8-10-r207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dorland. Dorland’s Illustrated Medical Dictionary. Amsterdam, Netherlands: Saunders (Elsevier); 2007. [Google Scholar]
- 15.Merriam-Webster. Webster's Third New International Dictionary. Massachusetts: Merriam Webster Inc, Springfield; 2000. [Google Scholar]
- 16.Kogenaru S, del Val C, Hotz-Wagenblatt A, Glatting K-H. TissueDistributionDBs: a repository of organism-specific tissue-distribution profiles. Theor. Chem. Acc. 2010;125:651–658. [Google Scholar]
- 17.Vizcaino JA, Côté R, Reisinger F, Foster JM, Mueller M, Rameseder J, Hermjakob H, Martens L. A guide to the Proteomics Identifications Database proteomics data repository. Proteomics. 2009;9:4276–4283. doi: 10.1002/pmic.200900402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mosca E, Alfieri R, Merelli I, Viti F, Calabria A, Milanesi L. A multilevel data integration resource for breast cancer study. BMC Syst. Biol. 2010;4:76. doi: 10.1186/1752-0509-4-76. [DOI] [PMC free article] [PubMed] [Google Scholar]