Abstract
Consideration of biomolecules in terms of their molecular building blocks provides valuable new information regarding their synthesis, degradation and similarity. Here, we present the FragmentStore, a resource for the comparison of fragments found in metabolites, drugs or toxic compounds. Starting from 13 000 metabolites, 16 000 drugs and 2200 toxic compounds we generated 35 000 different building blocks (fragments), which are not only relevant to their biosynthesis and degradation but also provide important information regarding side-effects and toxicity. The FragmentStore provides a variety of search options such as 2D structure, molecular weight, rotatable bonds, etc. Various analysis tools have been implemented including the calculation of amino acid preferences of fragments’ binding sites, classification of fragments based on the enzyme classification class of the enzyme(s) they bind to and small molecule library generation via a fragment-assembler tool. Using the FragmentStore, it is now possible to identify the common fragments of different classes of molecules and generate hypotheses about the effects of such intersections. For instance, the co-occurrence of fragments in different drugs may indicate similar targets and possible off-target interactions whereas the co-occurrence of fragments in a drug and a toxic compound/metabolite could be indicative of side-effects. The database is publicly available at: http://bioinformatics.charite.de/fragment_store.
INTRODUCTION
Across all kingdoms of life, biomolecules are formed from molecular building blocks, suggesting that this principle has been favoured during evolution. During metabolism, the building block nature of biomolecules facilitates their degradation into fragments. A systems biology view of metabolism would benefit from considering these fragments. For instance, a study investigating the set of metabolites available to different organisms found that common substructures were observed within the uniquely used compounds in metabolic pathways, indicating that there are metabolite-mediated relationships between different organism groups (1).
During the past few decades, relatively few drugs have reached the market due to high failure rates at the clinical testing stage (2). The two main causes of these failures are lack of efficacy and toxicity (3). In fact, one-third of potential therapeutic compounds fail in clinical trials or are removed from the market at a later stage due to unacceptable side-effects, often caused by the drug binding to an off-target (4). Such drug polypharmacology has driven the prediction and characterization of drug-target associations in order to identify possible side-effects of drugs and identify new opportunities for therapeutic intervention (5–8). Fragment-based drug design is a well-established approach that has led to the successful development of novel leads for many different targets (9). However, fragment-based approaches can also be employed to perform small molecule building-block analyses for the identification of fragments responsible for off-target binding and side-effects (10). The Biochemical Substructure Search Catalogue (BiSSCat) stores computationally constructed substructures of compounds and can be used to determine possible additional substrates for enzymes (11). Identification of fragments that have a role in toxicity, side-effects or that mediate off-target interactions would aid the development of safe and effective medical drugs. Furthermore, such analyses could help to improve future toxicity testing in line with the US National Academy of Sciences’ recommendations to increase efficiency and decrease animal usage (12).
Comparison of the fragments present in different classes of small molecules such as metabolites, toxic compounds and drugs facilitates the answering of questions such as: (i) how many common fragments are there in the different classes of molecules? (ii) How does the synthesis and/or degradation of metabolites depend on their fragment composition? (iii) Do those molecules containing toxic fragments cause more side-effects? (iv) Can knowledge about common fragments in small molecules help optimize drug polypharmacology? (v) Can side-effects of drugs be rationalized through the identification of common fragments with metabolites? To help researchers answer such questions we developed the FragmentStore database which consists of more than 35 000 fragments and property data such as physicochemical information and binding site preferences.
THE DATABASE
The FragmentStore database consists of more than 35 000 different fragments resulting from fragmentation of more than 13 000 metabolic compounds, 2200 toxic compounds and 16 000 drugs and pharmacologically characterized compounds using two different fragmentation strategies: (i) the compounds were recursively fragmented according to the recap-rules and (ii) chains between ring structures were cut out.
For completeness, the compounds were also recursively fragmented according to their rotatable bonds, which alone resulted in more than 150 000 fragments. Properties such as molecular weight, logP and hydrophobicity are stored for all fragments. Furthermore, binding site preferences were determined for each fragment using all structures in the Protein Data Bank (PDB) (13) bound to a ligand of which the fragment is part of (if at least one crystallized structure is available in the PDB). These binding site preferences are calculated based on the frequency of amino acid types binding a fragment compared to the amino acid frequencies for the entire protein surface. The amino acid binding site preferences for each fragment are displayed in a histogram with one bar for each amino acid, thus allowing users to ascertain whether there are particular patterns of amino acids responsible for binding particular fragments. Moreover, identical fragments in different binding sites are superimposed and shown with the amino acids that form the binding pocket. These superimpositions provide detailed information about the mechanism of fragment binding and provide valuable information about the specificity of interaction in both homologous and non-homologous proteins.
FragmentStore offers various ways of searching the database:
Fragments can be selected according to the rule used for fragmentation, based on their chemical properties or class.
It is also possible to search for fragments with a specific binding site amino acid or physicochemical composition.
An enzyme classification (EC) tree can be browsed and fragments which bind to a particular class of enzymes can be selected.
A SCOP (14) classification tree can be browsed and fragments which bind to a particular SCOP class of proteins can be selected.
Structural searches can be performed by uploading or drawing a molecule.
Property searches e.g. molecular weight, number of atoms etc. can be carried out.
Fragments can be searched using compound names (excluding those obtained from commercial databases).
A search box is available, called Fraggle, which allows users to enter text for searching against drug names and PDB header entries.
We have also implemented a fragment-assembler tool, which allows users to build a library of small molecules based on the selection of fragments of their choice using reverse recap-rules.
MATERIAL AND METHODS
Datasets
More than 35 000 fragments were generated by fragmenting three different compound libraries comprising metabolites, toxic molecules and drugs. More than 13 000 KEGG-metabolites (15) were fragmented for the metabolite dataset and more than 2200 compounds from the SuperToxic database (16) were fragmented for the toxic dataset.
Altogether, the drug dataset consists of fragments from more than 16 000 unique drugs from the following resources: SuperDrug (∼2400 drugs) (16), KEGG-drugs (∼7000) (15,17), DrugBank’s approved drugs (∼1300) (18), WDI drugs (Derwent World Drug Index) (∼7000) and CMC drugs (MDL) (∼8000). For the last two databases, we consider only drugs, which are publicly available, e.g. in PubChem. The fragments have direct links to the compounds of SuperDrug, KEGG-drugs, DrugBank, SuperToxic and KEGG-metabolites.
Fragmentation methods
The ligands in the above-mentioned datasets were fragmented using three different strategies. For the first strategy, the compounds were fragmented recursively using the recap-rules (19). The recap methodology helps to identify fragments which are useful for combinatorial chemistry. This fragmentation method allows libraries to be generated which contain fragments that can be easily connected by bonds that are easy to synthesize e.g. ester bonds. Altogether, the recap-rules comprise eleven different bonds: amide, ester, amine, urea, ether, olefin, quaternary nitrogen, bond between aromatic nitrogen and carbon, lactam-nitrogen and carbon, bonds between aromatic rings and sulphonamide.
For the second strategy, chains between two ring structures are cut out. Due to its non-redundancy, this fragment library is more suitable for statistical analysis than libraries generated using recursive methods. For the third strategy, the ligands were recursively fragmented by their rotatable bonds. The latter fragmentation rule produced the most fragments (see Figure 1).
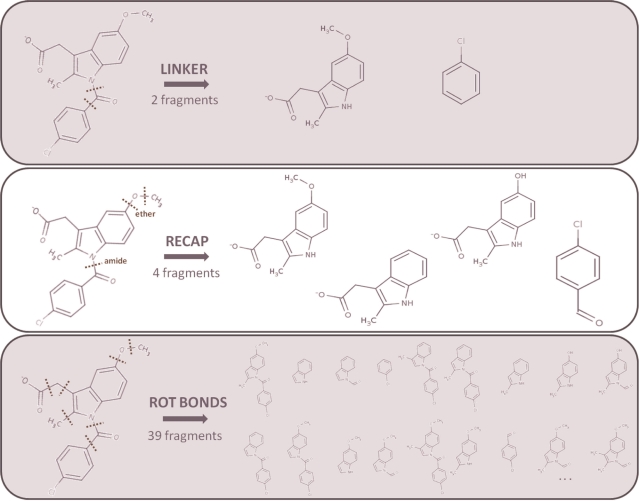
Figure 1.
Three methods of fragmentation. The figure shows only the fragments which break no more than one of the rules of the rule-of-three. In the top panel, a compound is fragmented according to the linker rule, producing two fragments. The middle panel shows the same compound being fragmented according to the recap-rules, producing a total of four fragments. Here, two other generated fragments were excluded because they broke more than one rule (fragments not shown). The bottom panel shows the same compound being fragmented according to its rotatable bonds, producing a total of 39 fragments (only a subset is shown for clarity). Thirteen other fragments were excluded using the rule-of-three.
To validate the fragments for inclusion into FragmentStore, a modification of the Lipinski rule-of-five was used. Astex Technology’s rule-of-three is useful for constructing fragment libraries that are efficient for lead generation (20). The rule-of-three criteria for fragments (which can later be combined into compounds) are that they should have a molecular weight no more than 300 g/mol and that the number of hydrogen bond donors, the number of hydrogen acceptors and the clogP-value should not be more than three. Additionally, two properties should also be considered during the selection of fragments for building the fragment library: the number of rotatable bonds has to be less than four and the polar surface area has to be at most 60. For inclusion into the FragmentStore database, the fragments are only allowed to break one of these rules. Furthermore, every fragment in the FragmentStore consists of at least three heavy atoms.
Calculation of the binding site properties
After fragmentation, the binding site preferences for the fragments were calculated for all fragments which are co-crystallized in the Protein Data Bank. For each amino acid, the frequency of occurrence is calculated at the fragment’s binding site and compared to the frequency of occurrence at the protein’s surface. The binding site of a fragment is defined as all amino acids within 5 Å of the fragment. FragmentStore provides these binding site preferences as bar charts. The fragments which were co-crystallized in more than one different protein structure were superimposed using the superimposition function of PyMOL (21). The superimposed fragments and its binding sites are visualized using Jmol.
Fragment-assembler
We have also implemented a fragment-assembler tool, which allows users to build a library of small molecules based on the selection of fragments of their choice using reverse recap-rules. The user is allowed to choose up to three fragments, which are combined to make new compounds that satisfy Lipinski’s rule-of-five (22). This rule defines properties, which compounds should fulfil to become drug candidates. This rule claims that an orally available drug has no more than five hydrogen bond donors, no more than ten hydrogen bond acceptors, a molecular weight of <500 g/mol and the LogP-value, which gives information about the lipophilicity of a molecule and is defined as the logarithm of the 1-octanol/water partition, should be below five.
As fragment assembly is computationally expensive, the user is sent the results (in SMILES format) by email within 20 min. The FragmentStore also provides an example set of fragments that can be used to demonstrate the capabilities of the fragment-assembler.
Structural fingerprint and similarity search
In order to search for fragments using structural features, bit vector `structural fingerprints’, which encode chemical and topological characteristics of a molecule, were included. The structural fingerprint was implemented using Open Babel (http://openbabel.sourceforge.net/), which offers four different fingerprints (FP2, FP3, FP4, MACCS).
Fingerprint 2 (http://openbabel.org/wiki/FP2) is widely used for the comparison of small molecules and is path- based and indexes linear fragments up to seven atoms. However, this fingerprint is not optimal for the comparison of small fragments. To provide an optimal comparison of small fragments, a combination of fingerprint 2 and 4 (FP2, FP4) was used. Fingerprint 4 (http://openbabel.org/wiki/FP4) is based on a set of SMARTS patterns and also considers functional groups. The combined fingerprint shows the best results in comparing fragments.
This combined structural fingerprint is pre-calculated for all fragments in the database and will be calculated for the query fragments to compare it to the entries of FragmentStore. For the similarity search the Tanimoto coefficient is used, which gives values in the range of zero (no bits in common) to unity (all bits the same).
Server
FragmentStore is designed as a relational database on a MySQL server. Additionally, the MyChem package (http://mychem.sourceforge.net/) is installed to provide a complete set of functions for handling chemical data within MySQL. Most of the functions used by MyChem depend upon Open Babel. The structural fingerprint is implemented in Open Babel 2.2.3 (http://openbabel.sourceforge.net/). To allow the upload or drawing of a query structure, the Marvin Sketch plugin (http://www.chemaxon.com) was installed. For the visualisation of the 3D structures Jmol (http://www.jmol.org/) was installed. The website is built with php and web access is enabled via Apache HTTP Server 2.2.
EXAMPLE OF USE
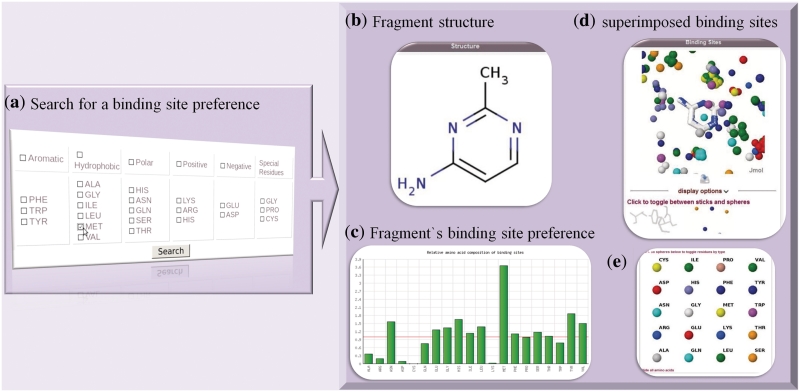
If one wants to find a ligand for a specific binding site of a target, the first step could be the characterisation of the pocket. Afterwards, fragments for the specific binding site can be detected in FragmentStore. If, for example, one part of the binding site consists of many hydrophobic amino acids like methionine, the user is able to search the FragmentStore database for fragments which have hydrophobic binding site preferences. Beside the fragment and its physicochemical properties, the user gets the binding site preference as a bar plot. Furthermore, the binding sites in which the fragment occurs are superimposed and displayed in 3D using Jmol (Figure 2).
Figure 2.
The binding site search feature of the FragmentStore can be used to retrieve fragments according to the amino acid type or physicochemical properties of the residues they bind to. (a) First, the user selects the particular amino acids that they are interested in. In this case all fragments that are crystallized in a binding pocket containing methionine residues will be retrieved. The results returned include: (b) a 2D structure of the fragment; (c) the binding site amino acid propensities; (d) a Jmol applet displaying the superimposed binding sites and (e) a key for the different amino acids which can be switched on and off in the applet.
FIRST ANALYSIS
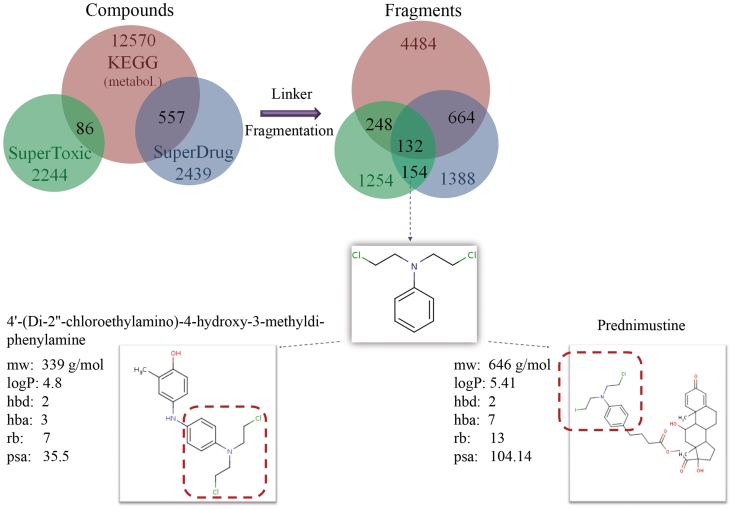
In the following analysis we consider the fragments which are produced after fragmenting the SuperDrug drugs, the KEGG-metabolites and the highly toxic compounds from the SuperToxic database using the linker strategy. All ligands common to the SuperDrug and SuperToxic datasets were excluded from the latter dataset and all ligands with e.g. a ‘R’- or ‘*’-atom were excluded from the KEGG-metabolites. The intersection of the three ligand and resulting fragment datasets are shown in Figure 3. Only a small number of fragments are shared between all three classes of molecules. These fragments tend to be very small and are probably not essential for the compound’s specificity. As one would expect, there are proportionally less fragments shared between the toxic and metabolite fragments in comparison to those shared between the drugs and metabolite fragments. Surprisingly, although the toxic and drug datasets have no common compounds, the datasets share many similar fragments. These may contribute to the toxic effect of drugs and even side-effects. Figure 3 shows an example of a fragment which only occurs in the toxic and drug dataset but not in the KEGG-metabolites. The fragment is part of the chemotherapeutic drug, Prednimustine (23) and of several toxic compounds, e.g. 4′-(di-2″-chloroethylamino)-4-hydroxy-3-methyldiphenylamine. The compound 4′-(di-2″-chloroethylamino)-4-hydroxy-3-methyldiphenylamine was shown to have an LD50 value of 1.43 mg/kg (i.p.) in rat (24) and is therefore highly toxic.
Figure 3.
The intersection between metabolite, toxic and SuperDrug compounds compared to the intersection of their respective fragments after linker fragmentation. Only a small proportion of fragments are shared between all three classes. Although the toxic and drug dataset have no common compounds, the datasets share many similar fragments; these common fragments may contribute to drug toxicity and may even have a role in side-effects of medications. One such fragment, which is found in both the toxic and drug fragment dataset is shown. This is part of the chemotherapeutic drug, Prednimustine and of the toxic compound 4′-(di-2″-chloroethylamino)-4-hydroxy-3-methyldiphenylamine. mw: molecular weight; hbd: hydrogen bond donors; hba: hydrogen bond acceptors; rb: rotateable bonds; psa: polar surface area.
CONCLUSION AND FUTURE DIRECTIONS
The FragmentStore provides data on fragments from drugs, metabolites and toxic compounds. A fragmentation method should consider synthetic rules and distinguish between linkers and fragments. Co-occurrence of fragments in different drugs may indicate similar (off–) targets and the co-occurrence of fragments in drugs and toxic compounds or metabolites could be indicative for side effects.
The systematic (computational) synthesis of libraries from three fragments, as provided by the fragment-assembler in FragmentStore, leads on average to 10 000 compounds, which would be reasonable to sample the chemical space of a particular medical target.
A future goal of the FragmentStore is a mapping of all fragments onto metabolic and signaling pathways, hopefully elucidating interrelations between fragments, drugs, targets and therapeutic effects. For the mapping we will consider subtle changes and stereochemistry between the enzymatic steps of metabolic pathways. In a next step in-depth analysis will be carried out regarding the compounds acting on different receptors in the signaling cascades. The result will be a distribution of fragments/scaffolds over certain regions of regulation—such as particular kinases or neuronal receptors that might explain effects like multi-specificity.
AVAILABILITY
The FragmentStore database is freely available under the URL: http://bioinformatics.charite.de/fragment_store/ and will be updated regularly.
FUNDING
This work was supported by Deutsche Forschungsgemeinschaft (SFB 449), International Research Training Group (IRTG) Berlin–Boston–Kyoto, Bundesministerium für Bildung und Forschung (BMBF) and European Union (EU). Funding for open access charge: DFG, BMBF and EU.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank B. Grüning for help with MyChem and database setup and U. Schmidt for her support with the toxicity data and the figures. They would also like to thank A. Chefai for her support in developing the fragment-assembler.
REFERENCES
- 1.Muto A, Hattori M, Kanehisa M. Analysis of common substructures of metabolic compounds within the different organism groups. Genome Inform. 2007;18:299–307. [PubMed] [Google Scholar]
- 2.Enna SJ, Williams M. The decreased number of new drug approvals (NDAs) has been a topic of considerable debate over the past decade. Preface. Adv. Pharmacol. 2009;57:xi–ii. doi: 10.1016/S1054-3589(08)57015-3. [DOI] [PubMed] [Google Scholar]
- 3.Kola I, Landis J. Can the pharmaceutical industry reduce attrition rates? Nat. Rev. Drug Discov. 2004;3:711–715. doi: 10.1038/nrd1470. [DOI] [PubMed] [Google Scholar]
- 4.Kennedy T. Managing the drug discovery/development interface. Drug Discov. Today. 1997;2:436–444. [Google Scholar]
- 5.Bajorath J. Computational analysis of ligand relationships within target families. Curr. Opin. Chem. Biol. 2008;12:352–358. doi: 10.1016/j.cbpa.2008.01.044. [DOI] [PubMed] [Google Scholar]
- 6.Chen B, Wild D, Guha R. PubChem as a source of polypharmacology. J. Chem. Inf. Model. 2009;49:2044–2055. doi: 10.1021/ci9001876. [DOI] [PubMed] [Google Scholar]
- 7.Keiser MJ, Setola V, Irwin JJ, Laggner C, Abbas AI, Hufeisen SJ, Jensen NH, Kuijer MB, Matos RC, Tran TB, et al. Predicting new molecular targets for known drugs. Nature. 2009;462:175–181. doi: 10.1038/nature08506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kuhn M, Campillos M, Gonzalez P, Jensen LJ, Bork P. Large-scale prediction of drug-target relationships. FEBS Lett. 2008;582:1283–1290. doi: 10.1016/j.febslet.2008.02.024. [DOI] [PubMed] [Google Scholar]
- 9.Hajduk PJ, Greer J. A decade of fragment-based drug design: strategic advances and lessons learned. Nat. Rev. Drug Discov. 2007;6:211–219. doi: 10.1038/nrd2220. [DOI] [PubMed] [Google Scholar]
- 10.Siegel MG, Vieth M. Drugs in other drugs: a new look at drugs as fragments. Drug Discov. Today. 2007;12:71–79. doi: 10.1016/j.drudis.2006.11.011. [DOI] [PubMed] [Google Scholar]
- 11.Kotera M, McDonald AG, Boyce S, Tipton KF. Functional group and substructure searching as a tool in metabolomics. PLoS One. 2008;3:e1537. doi: 10.1371/journal.pone.0001537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Committee on Toxicity Testing and Assessment of Environmental Agents, National Research Council of The National Academies. Toxicity Testing in the 21st Century: A Vision and A Strategy. Washington, DC: The National Academies Press; 2007. [Google Scholar]
- 13.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lo Conte L, Ailey B, Hubbard TJ, Brenner SE, Murzin AG, Chothia C. SCOP: a structural classification of proteins database. Nucleic Acids Res. 2000;28:257–259. doi: 10.1093/nar/28.1.257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kanehisa M. The KEGG database. Novartis Found. Symp. 2002;247:91–101; discussion 101–103, 119–128, 244–152. [PubMed] [Google Scholar]
- 16.Schmidt U, Struck S, Gruening B, Hossbach J, Jaeger IS, Parol R, Lindequist U, Teuscher E, Preissner R. SuperToxic: a comprehensive database of toxic compounds. Nucleic Acids Res. 2009;37:D295–D299. doi: 10.1093/nar/gkn850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kanehisa M, Goto S, Furumichi M, Tanabe M, Hirakawa M. KEGG for representation and analysis of molecular networks involving diseases and drugs. Nucleic Acids Res. 2010;38:D355–D360. doi: 10.1093/nar/gkp896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wishart DS, Knox C, Guo AC, Cheng D, Shrivastava S, Tzur D, Gautam B, Hassanali M. DrugBank: a knowledgebase for drugs, drug actions and drug targets. Nucleic Acids Res. 2008;36:D901–D906. doi: 10.1093/nar/gkm958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lewell XQ, Judd DB, Watson SP, Hann MM. RECAP – retrosynthetic combinatorial analysis procedure: a powerful new technique for identifying privileged molecular fragments with useful applications in combinatorial chemistry. J. Chem. Inf. Comput. Sci. 1998;38:511–522. doi: 10.1021/ci970429i. [DOI] [PubMed] [Google Scholar]
- 20.Congreve M, Carr R, Murray C, Jhoti H. A ‘rule of three' for fragment-based lead discovery? Drug Discov. Today. 2003;8:876–877. doi: 10.1016/s1359-6446(03)02831-9. [DOI] [PubMed] [Google Scholar]
- 21.DeLano WL. The PyMOL Molecular Graphics System. LLC: Schrödinger; [Google Scholar]
- 22.Lipinski CA. Chris Lipinski discusses life and chemistry after the Rule of Five. Drug Discov. Today. 2003;8:12–16. doi: 10.1016/s1359-6446(02)02556-4. [DOI] [PubMed] [Google Scholar]
- 23.Unterhalt M, Herrmann R, Tiemann M, Parwaresch R, Stein H, Trumper L, Nahler M, Reuss-Borst M, Tirier C, Neubauer A, et al. Prednimustine, mitoxantrone (PmM) vs cyclophosphamide, vincristine, prednisone (COP) for the treatment of advanced low-grade non-Hodgkin's lymphoma. German Low-Grade Lymphoma Study Group. Leukemia. 1996;10:836–843. [PubMed] [Google Scholar]
- 24.Ross WC. Aryl-2-Halogenoalkylarylamines. Xxi. The design of agents to exploit differences in cellular oxidation-reduction potentials. Biochem. Pharmacol. 1964;13:969–982. doi: 10.1016/0006-2952(64)90093-0. [DOI] [PubMed] [Google Scholar]