Abstract
We present WebGeSTer DB, the largest database of intrinsic transcription terminators (http://pallab.serc.iisc.ernet.in/gester). The database comprises of a million terminators identified in 1060 bacterial genome sequences and 798 plasmids. Users can obtain both graphic and tabular results on putative terminators based on default or user-defined parameters. The results are arranged in different tiers to facilitate retrieval, as per the specific requirements. An interactive map has been incorporated to visualize the distribution of terminators across the whole genome. Analysis of the results, both at the whole-genome level and with respect to terminators downstream of specific genes, offers insight into the prevalence of canonical and non-canonical terminators across different phyla. The data in the database reinforce the paradigm that intrinsic termination is a conserved and efficient regulatory mechanism in bacteria. Our database is freely accessible.
INTRODUCTION
Transcription termination is an important regulatory step of gene expression. All RNA polymerases that transcribe a DNA template must terminate, dissociate and release the product RNA at a defined position or region on the DNA. The RNA structure involved in this process is called a tanscrtiption terminator (1–3). In bacteria, wherein detailed studies have been carried out, termination is achieved by two mechanisms—intrinsic (factor independent) and factor dependent. The former process is primarily dependent on the secondary structure formed in the nascent RNA and can function in a minimal in vitro system in the absence of other proteins factors (4–6). In contrast, factor-dependent termination relies on proteins such as Rho and the Nus factors (7,8).
Once formed during transcription, the terminator interacts with RNA polymerase resulting in destabilization and dissociation of the ternary elongation complex (TEC) (3,9–11). Based on the studies in Escherichia coli, an intrinsic terminator is a RNA structure consisting of a guanidine-cytidine content (GC)-rich hairpin immediately followed by a stretch of 6–8 U residues. Although such terminators were found in many genomes, their occurrence is rare in several other genomes when the stringent parameters were applied for the analysis. With the development of newer algorithms which could analyse genomes with different criteria, variant (non-canonical) terminators were detected and experimentally verified (12–20). Indeed, since intrinsic termination is an ancient and conserved mechanism, it is not surprising that all bacteria rely on this regulatory mechanism.
The exponential increase in available genomic data has now allowed us to analyse and catalogue the terminator content of nearly 2000 sequences (chromosomal and plasmid) of bacterial origin. Here, we present WebGeSTer DB (http://pallab.serc.iisc.ernet.in/gester), the largest collection of intrinsic terminators from all completely sequenced bacterial genomes and plasmids. The database has been compiled using WebGeSTer, an improved version of GeSTer (20). At present, WebGeSTer DB consists of all types of intrinsic terminators identified in 1060 bacterial chromosomes and 798 plasmids available at the NCBI database (Table 1). In all, information about 977 173 terminators, both canonical and non-canonical, have been compiled in the database (Table 1). The terminator profile for whole genomes as well as for individual genes can be extracted from WebGeSTer DB. The occurrence of terminators with respect to specific genes can be visualized in a high-resolution map. Furthermore, the database has a user-friendly and interactive interface that allows investigators to obtain results in both graphic and tabular form. The parameters for terminator search can be user-defined and one can upload new genome sequences in FAST Alignment (FASTA) or GenBank format for analysis.
Table 1.
Summary of information available at WebGeSTer DB
| Bacterial genomes | 1060 |
| Plasmids | 798 |
| Genes | 3 335 043 |
| ‘All’ terminators | 1 228 606 |
| ‘Best’ terminators | 977 173 |
| L-shaped terminators | 523 243 |
| I-shaped terminators | 453 930 |
| U-shaped terminators | 100 364 |
| V-shaped terminators | 1615 |
| X-shaped terminators | 55 872 |
GENERATION OF DATABASE
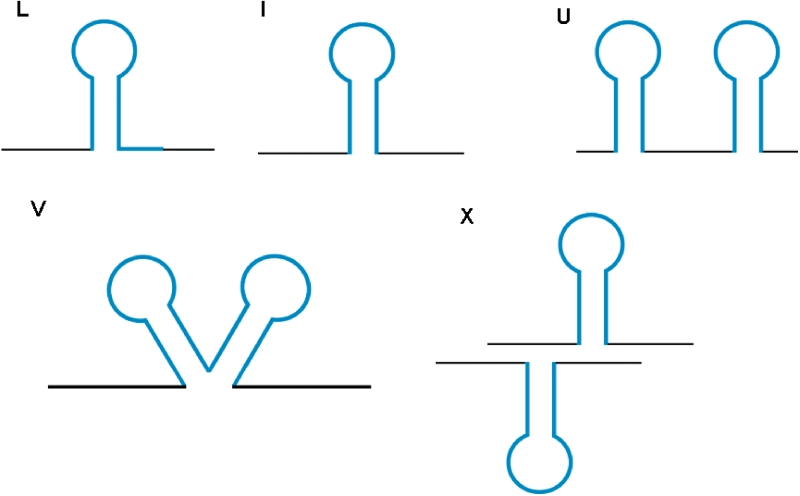
The terminator database has been compiled using WebGeSTer, developed from the parent program, GeSTer (20), incorporating several improvements. The details of WebGeSTer generation are available in the website. Briefly, WebGeSTer accepts sequences in both GenBank and FASTA format, extracts the regions of −20 to +270 bp relative to the stop codons from genomic sequences, and searches them for potential palindromic sequences. For the region downstream of every stop codon, all possible hairpins are computed and the most stable structure (with the most negative ΔG value) is selected as the ‘Best’ terminator. A genomic ΔGcut-off selects the final set of identified terminators. For any terminator, the sequence, genomic coordinates, structural parameters such as length of stem, loop, sequence following the hairpin mismatches and gaps can be obtained from the output. WebGeSTer can identify both canonical and non-canonical terminators and group them. The different types of terminators (Figure 1) catalogued in the WebGeSTer DB are:(i) L-shaped (canonical terminators): where the hairpin is followed by a 10 bp trail having >3 uridylates. The four types of non-canonical terminators are: (ii) I-shaped: where there are ≤3 uridines in the trail following the hairpin, (iii) U-shaped: when there is more than one hairpin structure in tandem with an interval of <50 nt between them, (iv) X-shaped: convergent type structures that function as terminators for the convergently transcribed genes on two different strands and (v) V-shaped: two hairpins, with the second hairpin starting immediately at the end of first one. In case of the U, V and X terminators, the individual structures can be L- or I-shaped. The program is adaptable in which the user can change the parameters such as stem-length, loop size, maximum allowance for mismatch and gap and also search region. The core algorithm of WebGeSTer was written in PERL and produced ASCII text files. From these files, data were extracted to populate the MySQL tables. The database was built using MySQL version 5.0.84 and interfaced using PERL version 5.10.0 and PHP version 5.2.9. The figures were drawn using GD library, version 2.45.To evaluate the accuracy and sensitivity of the WebGeSTer DB, a sample of 100 experimentally known terminators was assessed (14). The algorithm identified 91 of these terminators (Supplementary Table S1) and hence false negatives make up <10% of all the predictions. The detection ability of WebGeSTer was tested by drawing an receiver operating characteristic (ROC) curve for each genome. The ROC curves plot the probability of detection against the probability of false alarm at various input thresholds to the algorithm (16). The results for individual genomes are obtainable from the webpage. Further the validation of the predictions comes from analysing experimentally characterized operons (e.g. rrn, trp, thr, his operons of E.coli K-12). For these operons, WebGeSTer correctly predicts the terminators present at 3′ end, but not any ‘false’ intra-operonic terminators.
Figure 1.
Terminators catalogued in the WebGeSTer DB—(i) L-shaped (canonical terminators): hairpin + 10 bp trail having >3 uridylates, (ii) I-shaped: hairpin + ≤3 uridines in the trail, (iii) U-shaped: >1 hairpin in tandem with an interval of <50 nt in between, (iv) V-shaped: two hairpins’ structure, where the second structure starts immediately at the end of first one and (v) X-shaped: convergent type structures that function as terminators for the convergently transcribed genes on two different strands.
CONTENT AND INFORMATION RETRIEVAL
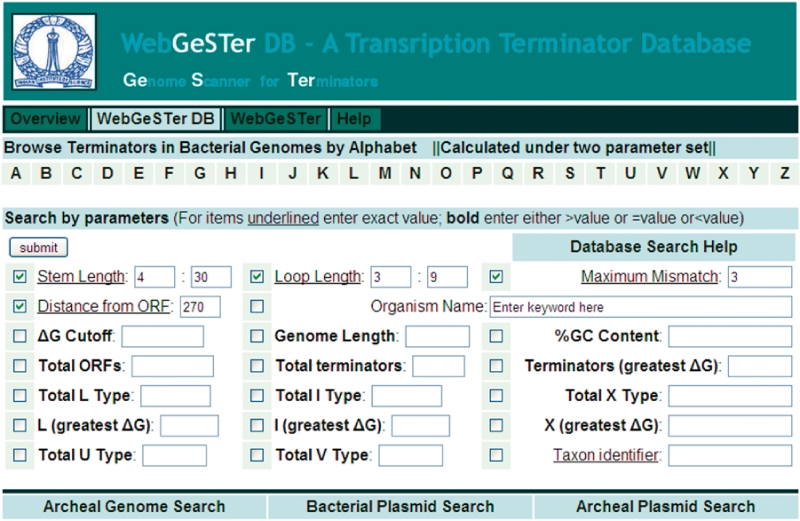
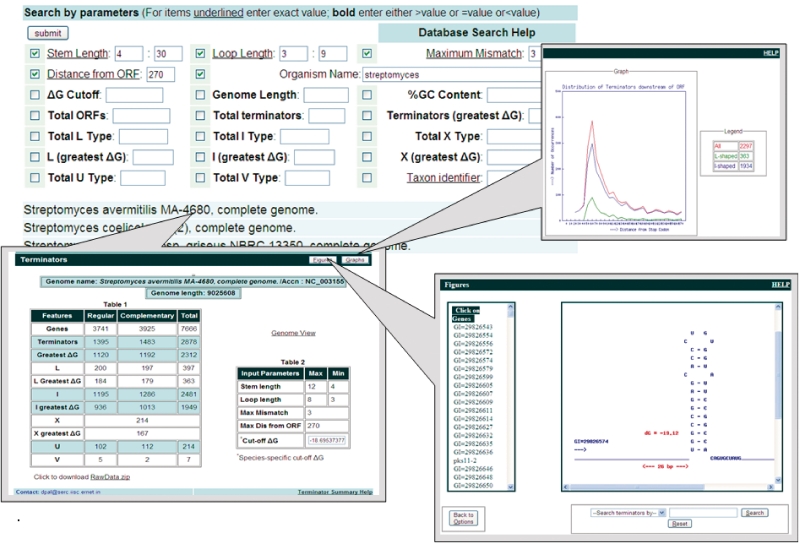
WebGeSTer DB is the largest compilation of intrinsic terminators till date. To facilitate retrieval of data from the WebGeSTer DB, the information has been arranged in different tiers ranging from different phyla to genomes of individual strains (Figure 2 and Table 2). A search initiated at a phylum (e.g. Firmicutes, Proteobacteria) can be threaded to finally reach the details of a particular terminator downstream of a specific gene-of-interest (Figure 3). In the database, users can find the terminator profiles of either an individual genome or all the member species of a given phylum/class by a easy-to-use search module. Information on a given genome has been further subdivided into files, which provide details, for e.g. of ‘All’ candidate terminators, the ‘Best’ candidate terminators and different types of terminators (L, I, U, V, X). The database contains several computed features for every individual terminator. These include its sequence, stem length, loop size, distance from gene, ΔG, etc. (Table 3). All the information can be downloaded as zipped files from the website for further processing.
Figure 2.
GeSTerDB and WebGeSTer interface. The user can refine his search for terminator profiles with one or more of the criteria provided. For e.g. a search for ‘Total ORFs >5000’ and ‘Terminators (lowest ΔG)>2500’ would retrieve all genomes which have more than 5000 genes and also greater than 2500 ‘Best’ terminators.
Table 2.
Salient searches at WebGeSTer DB
| Search by … | Example (type in … ) | Results |
|---|---|---|
| Organism | Mycobacterium tuberculosis | All M. tuberculosis strains and plasmids (if any) |
| Taxon | 1239 (taxon ID from NCBI) | All genomes belonging to phyla 1239, i.e. firmicutes |
| Stem length | 4:10 | Individual genomes which have terminators with stem length between 4 and 10 bp |
| GC content (%) | >60 | All genomes which have genomic GC content >60% |
| Total terminators | >4000 | All genomes with more than 4000 terminators |
| Terminators (greatest ΔG) and L (greatest ΔG) | >3000 and >2000 | All genomes which have >3000 ‘Best’ terminators, of which >2000 are L-shaped |
| Further searches | |
|---|---|
| From genome page … | Access to … |
| Download rawdata.zip | Individual files detailing genes, sequences, terminators, genomic coordinates… |
| Figures | Structures of all identified terminators- scroll down or search terminators by GI number, gene name … |
| Tables | Whole genome distribution of terminators—‘ALL’ palindromic structures, `Best' structures, L-shaped or I-shaped … |
| TER-MAP (genome browser for terminators) | High-resolution map of genome, with terminators (L or I-shaped) downstream of genes visible. By clicking on individual terminators, the user can gain access to its parameters |
Figure 3.
Progressive data accession in WebGeSTer DB. A search initiated at a specific genome can finally lead to details about a terminator downstream of a specific gene.
Table 3.
Parameters of terminators obtainable from WebGeSTer DB
| 1. | Length of stem of hairpin—subdivided into upstem length, downstem length |
| 2. | Sequence of stem—subdivided into upstem and downstem sequence |
| 3. | Length and sequence of mismatches and gaps |
| 4. | Size and sequence of loop |
| 5. | Distance of terminator from stop codon |
| 6. | ΔG of terminator |
| 7. | Accession number of gene |
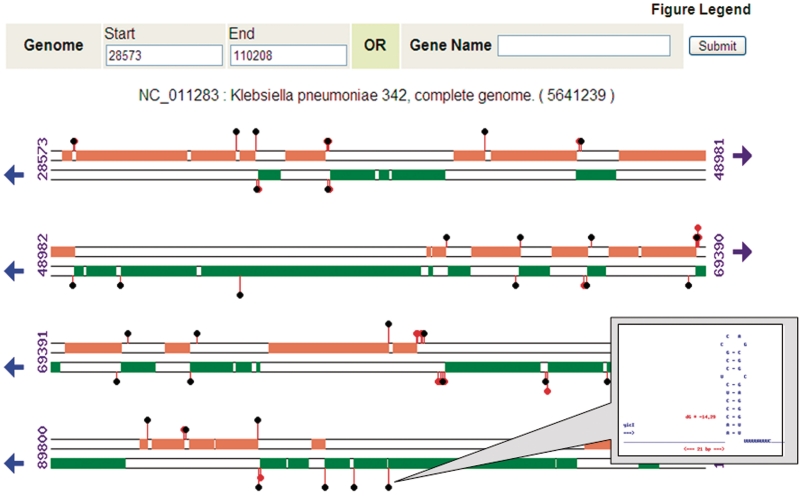
WebGeSTer DB also provides the user with whole-genome terminator maps. The genes and different types of terminators of any genomic region from all of the 1858 sequences (1060 chromosomes + 798 plasmids) can be visualized by TERminator MAP (TER-MAP), an interactive map at single gene level resolution (Figure 4). Genes and terminators of both strands are arranged in linear array in TER-MAP. ‘All’ identified palindromic structures are indicated and amongst them, the ‘Best’ terminator candidates are highlighted. Furthermore, the user can click onto the terminator-of-interest and be guided to the data for that specific terminator. Information about the genes can be similarly obtained that leads to the NCBI file (http://www.ncbi.nlm.nih.gov/protein) about that specific gene and gene product.
Figure 4.
TER-MAP—the high-resolution terminator map and browser. From the genome summary page, the user can navigate to a defined region of the genome or to a specific gene. Terminators are represented as ‘lollipops’ at ends of genes. Clicking on any terminator leads to more information about its computed structure and other parameters.
WebGeSTer works using a default set of parameters aimed to provide maximum number of accurate and sensitive predictions. These are: stem length between 4 and 30 bp, and loop size between 3 and 9 nt and maximum mismatch of 3 nt (12,14,15,19). However, experiments have suggested that an intrinsic terminator with a stem length of 8–9 bp is sufficient to enforce termination (10,21,22). Most experimentally known terminators have hairpins in this range. Furthermore, in silico analyses have previously shown that most terminators across diverse species have stem length between 6 and 13 bp (14,16–18). Keeping these results in consideration, two sets of data are present for each sequence in the database. One of them has been generated with the default settings of GeSTer (stem length between 4 and 30 bp, and loop size between 3 and 9 nt). For the second set, criteria for stem length was set at 4–12 bp, while loop size was 3–8 nt.
WebGeSTer DB is unique in housing information also on several types of non-canonical terminators. Experimentally, there is a substantial body of evidence for non-canonical terminators (I, U, X and V-shaped) in many species of mycobacteria, Streptomyces lividans and actinophages (12,13,19,20,23). Non-canonical terminators also occur at ends of several experimentally identified operons in diverse bacteria (17,18,24). Even the prototypical E. coli seems to have a large number of non-canonical terminators and experimentally a mutant E. coli RNA polymerase has been shown to terminate at such non-canonical terminators(25). Information about non-canonical terminators from GeSTer results has been applied to define operon boundaries in the S. coelicolor genome (26), a bacteria with few canonical terminators. Thus, by also compiling data about non-canonical terminators, WebGeSTer DB could be a starting point for further research into understanding the mechanism of termination and improving genome annotation. However, it is possible that a subset of hairpins identified is class I pause signals and not necessarily non-canonical terminators. No secondary structure prediction algorithm can distinguish between them. One would have to experimentally determine the 3′-end of the RNA to distinguish class I pause signals from the non-canonical terminators.
Earlier, GeSTer data has been used to find terminators in the archaea Thermococcus kodakarensis (27). WebGeSTer DB has now a collection of terminator profiles for 77 archaeal genomes and plasmids of archaeal origin. Archaea employ a different mechanism for transcription termination, which is dependent on presence of T-rich sequences downstream of the stop codon, that would get transcribed into a U-stretch in the transcript (27). Since all L-shaped terminators invariably consist of a U-trail, the program can also detect several such archaeal terminators.
Keeping in mind the new scenarios (e.g. meta genomes) where the WebGeSTer algorithm could be effective in detecting intrinsic terminators, we have upgraded WebGeSTer to accept FASTA sequences from external users (Supplementary Figure S1). This could also be particularly useful to researchers who need to analyse a sequence that has not yet been made available in the GenBank format. The database is freely accessible and will be updated on a regular basis.
ANALYSIS OF TERMINATORS ACROSS BACTERIA
WebGeSTer DB also houses a detailed analysis of the structural parameters of terminators, their prevalence and their divergence. The analysis was carried out using data from a large sample extracted from the database with representative species from 22 phyla. The salient findings are summarized below:
Intrinsic terminators are present in all bacterial genomes. Canonical or L-shaped terminators are the most abundant terminators (∼51% of ‘Best’ terminators). However, non-canonical terminators that have been experimental shown to be functional, are also present in large numbers (∼49%) (Supplementary Table S1).
Of the genes, 28.1% have a’ Best’ candidate terminator immediately downstream of its stop codon. Both canonical and non-canonical terminators tend to cluster within 50 bp of the stop codon in most species.
Substantial difference in terminator preference is observed across phyla. Some phyla show a preference for L-shaped terminators, while many others have larger representation of the I-shaped terminators (Supplementary Figure S2).
Across species, most terminators have a stem length of 7–14 bp and a loop size of 4 nt (Supplementary Figure S3). Since the ΔG of the terminator is mainly a function of its stem–loop structure, most of the identified terminators have ΔG in the range −15 to −25 kcal/mol (median value −18.1 kcal/mol)
The fraction of I-shaped terminators increases as the genomic GC content rises across phyla (Supplementary Figure S4). Thus, genomic GC content is one of the determinants of the type of terminator predominant in a given organism.
Transcription termination factor Rho is essential in many bacteria, while some other species do not have a rho gene. The terminator content of 55 bacterial genomes that lacked a rho gene was assessed and they have a preponderance of L-shaped terminators. Most of these bacteria belong to Firmicutes and Tenericutes.
CONCLUSIONS
WebGeSTer DB is a catalogue and presentation of intrinsic terminators. The data sets from WebGeSTer DB show that intrinsic termination is a universally conserved mechanism present in all bacterial species sequenced till date. The representative data from WebGeSTer DB are in agreement with the experimental evidence of intrinsic termination, and hence serve as a validation of the database. The database provides insight into the evolved variations in intrinsic terminators, like other successful regulatory process. The compilation would be invaluable for further experimentation on the mechanism of termination and understanding of gene expression in different bacteria.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
The work is supported by the Center for Excellence in Bioinformatics, Department of Biotechnology, Government of India and Center of Excellence for mycobacteria research grant, Department of Biotechnology, Government of India. Funding for open access charge: Center of Excellence for mycobacteria research, Department of Biotechnology, Government of India.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We thank Rupesh Kumar and Shyam Unniraman for the design of Figure 1 and discussions respectively. V.N. is a recipient of J.C.Bose fellowship of Department of Science and Technology, Government of India.
REFERENCES
- 1.von Hippel PH, Delagoutte E. A general model for nucleic acid helicases and their “coupling” within macromolecular machines. Cell. 2001;104:177–190. doi: 10.1016/s0092-8674(01)00203-3. [DOI] [PubMed] [Google Scholar]
- 2.Platt T. Transcription termination and the regulation of gene expression. Annu. Rev. Biochem. 1986;55:339–372. doi: 10.1146/annurev.bi.55.070186.002011. [DOI] [PubMed] [Google Scholar]
- 3.Borukhov S, Nudler E. RNA polymerase: the vehicle of transcription. Trends Microbiol. 2008;16:126–134. doi: 10.1016/j.tim.2007.12.006. [DOI] [PubMed] [Google Scholar]
- 4.Richardson JP, Greenblatt J. Control of RNA chain elongation and termination. In: Neidhert FC, editor. Escherichia coli and Salmonella: Cellular and Molecular Biology. 2nd Edn. Washington, DC: ASM press; 1996. pp. 822–848. [Google Scholar]
- 5.Henkin TM. Control of transcription termination in prokaryotes. Annu. Rev. Genet. 1996;30:35–57. doi: 10.1146/annurev.genet.30.1.35. [DOI] [PubMed] [Google Scholar]
- 6.Henkin TM. Transcription termination control in bacteria. Curr. Opin. Microbiol. 2000;3:149–153. doi: 10.1016/s1369-5274(00)00067-9. [DOI] [PubMed] [Google Scholar]
- 7.Richardson JP. Rho-dependent termination and ATPases in transcript termination. Biochim. Biophys. Acta. 2002;1577:251–260. doi: 10.1016/s0167-4781(02)00456-6. [DOI] [PubMed] [Google Scholar]
- 8.Banerjee S, Chalissery J, Bandey I, Sen R. Rho-dependent transcription termination: more questions than answers. J. Microbiol. 2006;44:11–22. [PMC free article] [PubMed] [Google Scholar]
- 9.Datta K, von Hippel PH. Direct spectroscopic study of reconstituted transcription complexes reveals that intrinsic termination is driven primarily by thermodynamic destabilization of the nucleic acid framework. J. Biol. Chem. 2008;283:3537–3549. doi: 10.1074/jbc.M707998200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Epshtein V, Cardinale CJ, Ruckenstein AE, Borukhov S, Nudler E. An allosteric path to transcription termination. Mol. Cell. 2007;28:991–1001. doi: 10.1016/j.molcel.2007.10.011. [DOI] [PubMed] [Google Scholar]
- 11.Artsimovitch I, Landick R. Pausing by bacterial RNA polymerase is mediated by mechanistically distinct classes of signals. Proc. Natl Acad. Sci. USA. 2000;97:7090–7095. doi: 10.1073/pnas.97.13.7090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ingham CJ, Hunter IS, Smith MC. Rho-independent terminators without 3′ poly-U tails from the early region of actinophage oC31. Nucleic Acids Res. 1995;23:370–376. doi: 10.1093/nar/23.3.370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Williams DL, Slayden RA, Amin A, Martinez AN, Pittman TL, Mira A, Mitra A, Nagaraja V, Morrison NE, Moraes M, et al. Implications of high level pseudogene transcription in Mycobacterium leprae. BMC Genomics. 2009;10:397. doi: 10.1186/1471-2164-10-397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.d'Aubenton Carafa Y, Brody E, Thermes C. Prediction of rho-independent Escherichia coli transcription terminators. A statistical analysis of their RNA stem-loop structures. J. Mol. Biol. 1990;216:835–858. doi: 10.1016/s0022-2836(99)80005-9. [DOI] [PubMed] [Google Scholar]
- 15.de Hoon MJ, Makita Y, Nakai K, Miyano S. Prediction of transcriptional terminators in Bacillus subtilis and related species. PLoS Comput. Biol. 2005;1:e25. doi: 10.1371/journal.pcbi.0010025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lesnik EA, Sampath R, Levene HB, Henderson TJ, McNeil JA, Ecker DJ. Prediction of rho-independent transcriptional terminators in Escherichia coli. Nucleic Acids Res. 2001;29:3583–3594. doi: 10.1093/nar/29.17.3583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Mitra A, Angamuthu K, Jayashree HV, Nagaraja V. Occurrence, divergence and evolution of intrinsic terminators across eubacteria. Genomics. 2009;94:110–116. doi: 10.1016/j.ygeno.2009.04.004. [DOI] [PubMed] [Google Scholar]
- 18.Mitra A, Angamuthu K, Nagaraja V. Genome-wide analysis of the intrinsic terminators of transcription across the genus Mycobacterium. Tuberculosis. 2008;88:566–575. doi: 10.1016/j.tube.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 19.Unniraman S, Prakash R, Nagaraja V. Alternate paradigm for intrinsic transcription termination in eubacteria. J. Biol. Chem. 2001;276:41850–41855. doi: 10.1074/jbc.M106252200. [DOI] [PubMed] [Google Scholar]
- 20.Unniraman S, Prakash R, Nagaraja V. Conserved economics of transcription termination in eubacteria. Nucleic Acids Res. 2002;30:675–684. doi: 10.1093/nar/30.3.675. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Gusarov I, Nudler E. The mechanism of intrinsic transcription termination. Mol. Cell. 1999;3:495–504. doi: 10.1016/s1097-2765(00)80477-3. [DOI] [PubMed] [Google Scholar]
- 22.Wilson KS, von Hippel PH. Transcription termination at intrinsic terminators: the role of the RNA hairpin. Proc. Natl Acad. Sci. USA. 1995;92:8793–8797. doi: 10.1073/pnas.92.19.8793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Pulido D, Jimenez A. Optimization of gene expression in Streptomyces lividans by a transcription terminator. Nucleic Acids Res. 1987;15:4227–4240. doi: 10.1093/nar/15.10.4227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Castillo AR, Arevalo SS, Woodruff AJ, Ottemann KM. Experimental analysis of Helicobacter pylori transcriptional terminators suggests this microbe uses both intrinsic and factor-dependent termination. Mol. Microbiol. 2008;67:155–170. doi: 10.1111/j.1365-2958.2007.06033.x. [DOI] [PubMed] [Google Scholar]
- 25.McDowell JC, Roberts JW, Jin DJ, Gross C. Determination of intrinsic transcription termination efficiency by RNA polymerase elongation rate. Science. 1994;266:822–825. doi: 10.1126/science.7526463. [DOI] [PubMed] [Google Scholar]
- 26.Laing E, Mersinias V, Smith CP, Hubbard SJ. Analysis of gene expression in operons of Streptomyces coelicolor. Genome Biol. 2006;7:R46. doi: 10.1186/gb-2006-7-6-r46. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Santangelo TJ, Cubonova L, Skinner KM, Reeve JN. Archaeal intrinsic transcription termination in vivo. J. Bacteriol. 2009;191:7102–7108. doi: 10.1128/JB.00982-09. [DOI] [PMC free article] [PubMed] [Google Scholar]