Abstract
High-resolution structures of proteins remain the most valuable source for understanding their function in the cell and provide leads for drug design. Since the availability of sufficient protein structures to tackle complex problems such as modeling backbone moves or docking remains a problem, alternative approaches using small, recurrent protein fragments have been employed. Here we present two databases that provide a vast resource for implementing such fragment-based strategies. The BriX database contains fragments from over 7000 non-homologous proteins from the Astral collection, segmented in lengths from 4 to 14 residues and clustered according to structural similarity, summing up to a content of 2 million fragments per length. To overcome the lack of loops classified in BriX, we constructed the Loop BriX database of non-regular structure elements, clustered according to end-to-end distance between the regular residues flanking the loop. Both databases are available online (http://brix.crg.es) and can be accessed through a user-friendly web-interface. For high-throughput queries a web-based API is provided, as well as full database downloads. In addition, two exciting applications are provided as online services: (i) user-submitted structures can be covered on the fly with BriX classes, representing putative structural variation throughout the protein and (ii) gaps or low-confidence regions in these structures can be bridged with matching fragments.
INTRODUCTION
Proteins are by far the most versatile and complex molecules in the cell. It is commonly accepted that protein function directly relates to three-dimensional (3D) structure. Yet, for just over a quarter of all single-domain protein families detailed structural information is available (1), a number that can be extended through threading and homology modeling (2). Due to experimental constraints of X-ray crystallography or NMR, the rate at which new structures are determined is considerably slower than the amount of new sequence data that is being determined by next-generation sequencing methods.
In order to understand the structural protein universe, proteins have been classified on the architecture of the fold and evolutionary relationships in databases such as SCOP (3) or CATH (4). However, proteins often perform their functions using just a limited number of residues, making it worthwhile to find structural similarities at the level of protein fragments. Seeking for a ‘parts list’ of proteins—with α-helices and β-sheets as prime examples of common parts—fragment libraries have been constructed based on the similarity of the polypeptide backbone (5,6). These protein fragment libraries have been widely used for a range of applications such as structural comparison of protein folds through a simplified representation with fragments (7), homology modeling at the level of fragments (8,9), investigating sequence-to-structure relationships (10), approximating tertiary structure of proteins using fragments (11–14), loop prediction (15–17) or even novel fold prediction (18,19).
Unfortunately, many of the available fragment libraries are either limited in fragment classes or ‘states’ (6,20) or not publicly accessible (13). Moreover, existing databases are often biased towards short stretches of residues, typically three to nine residues long, or contain an extensive parts list but are not clustered based on backbone similarity, thereby complicating comparative studies (21). Although limited alphabets have been shown to successfully reconstruct existing proteins to global fits of 0.5 Å root mean square distance (RMSD) or serve successfully as templates to efficiently sample the protein space, they are too limited to describe protein structure at sub-ångström resolution, especially in the case of loops (22). To overcome these limitations we have constructed BriX, a database of protein fragments from 4 to 14 residues, hierarchically clustered on backbone similarities (22).
Here we describe how we updated the BriX database, which previously contained fragments from 1259 structures, to incorporate over 7000 structures from the ASTRAL40 set (a curated set of proteins with <40% sequence homology) (23). Furthermore, we enriched the database with all loops from over 14 000 structures in the ASTRAL95 set (sharing <95% sequence homology) and clustered these loops in their own respect. We also provide a user-friendly web interface to explore both BriX and Loop BriX (http://brix.crg.es). Finally, to illustrate the potential of our database we allow users to upload their own PDB structure and ‘cover’ parts or ‘bridge’ gaps with BriX or Loop BriX fragments. The new release of BriX is expected to be helpful to the scientific community by facilitating the use of fragments in structural biology, protein modeling and design.
DATABASE CONTENTS
Update of the BriX database
The first version of the BriX database (22) was constructed from the Whatif set of 1259 non-redundant proteins (24). Using a sliding-window technique, we segmented all proteins into fragments of 4 to 14 residues long and clustered them on their backbone similarity with a hierarchical clustering algorithm. The similarity between two fragments is defined as the average RMSD between the backbone atoms (N, Cα, C, O) of each corresponding residue.
The updated version of the BriX database is enriched with the much larger ASTRAL40 set of 7290 proteins sharing <40% of sequence homology. The ASTRAL40 set is a complete representation of the variety present in structural databases such as SCOP (Supplementary Figure S1). Once more, we fragmented all proteins and assigned each fragment to the closest class represented by its centroid. As it turns out, we were able to fit most of the ASTRAL40 fragments into existing BriX classes, showing the completeness of our structural alphabet in the updated version of BriX, while increasing its content 7-fold (Figure 1).
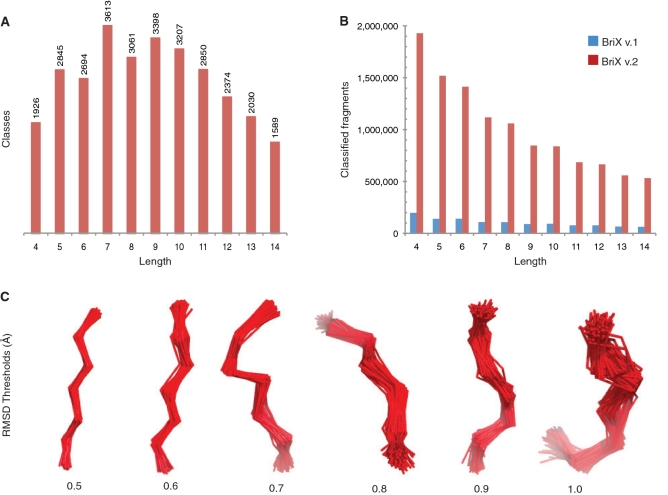
Figure 1.
The BriX database. (A) Number of BriX classes for lowest class thresholds per length. A peak in the number of classes can be observed at fragment length 7 and class threshold 0.5 Å. (B) Increase in the number of classified fragments from the first version of BriX (22) to the current version. (C) BriX classes with class thresholds varying from 0.5 to 1.0 Å RMSD for fragments of length 7. The class threshold indicates the compactness and structural homogeneity of the class, with lower thresholds causing classes to be more compact than higher thresholds.
BriX statistics
As expected, the number of classes varies with the length of the clustered fragments: even for short fragment length (n = 4) and strict threshold (≤0.4 Å RMSD) a large number of classes (2000) were observed. The largest amount of structural classes is detected when applying a clustering threshold of 0.5 Å to fragments of length 7: 3613 classes can be distinguished. Hereafter the number of classes steadily decreases until 1500 classes at length 14 (Figure 1A). As expected, the number of classes per length decreases with increasing classification thresholds (Supplementary Figure S2) as more different fragments are classified into a single class. Also, the percentage of classified fragments decreases steadily with increasing fragment length. To compensate for this, increasing the covering thresholds for a specific length improves the classification rates (Supplementary Figure S3).
Furthermore, we analyzed the secondary structure content in classes derived for different fragment lengths and thresholds. Not surprisingly, α-helical and β-strand fragments remain well represented in structural classes of higher length (Supplementary Figure S4), while loop fragments are under-represented in classes of all lengths, indicating that they are harder to classify. Clearly the majority of unclassified fragments are composed of loop structures (Supplementary Figure S5). This indicates that a separate classification scheme, more suited to the particularities of loop structures, could significantly enrich the BriX database.
Creation of the Loop BriX database
The Loop BriX database was built using 14 525 protein structures derived from the ASTRAL95 set containing protein structures sharing <95% sequence identity (23). A loop fragment starts and ends with a single residue belonging to a regular secondary structure such as a helix or a strand and contains any number of irregular residues in between. As shown by different studies, the structural loop space can be partitioned by four combinations of flanking regular elements: α-α, α-β, β-α and β-β Added proper references at the reference section (25–27) (Supplementary Figure S6).
We have introduced a novel way to compare the similarity between two loop fragments based on the (i) the distance between their end points (‘end-to-end distance’) rather than the overall structure similarity used in BriX and (ii) the superposition of two regular anchor residues at each side of the loop with a RMSD <1 Å. First, loops in each of the four loop classes described above were clustered on end-to-end distance using the same hierarchical clustering algorithm. These ‘super classes’ are composed of varying sizes and thus show a considerable amount of variation in the part between the end points (Figure 2A). Secondly, super classes were clustered in ‘sub classes’, grouping loops of the same length and similar structure.
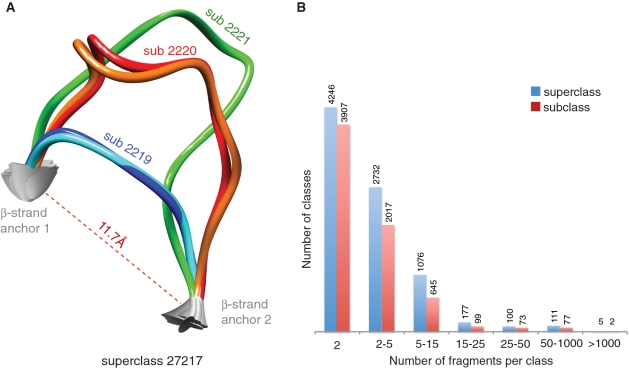
Figure 2.
The Loop BriX database. (A) Example of a superclass containing three subclasses. The superclass contains fragments with end-to-end distance around 11.78 Å RMSD and two β-strand anchor residues. At the subclass level, fragments with similar length and backbone are grouped (length 7 for subclass 2219 and length 13 for subclass 2220 and 2221, superposition threshold of 1 Å). (B) Number of superclasses (blue) and subclasses (red) per class size, distributed in bins. In general, classes from Loop BriX are less populated than classes from BriX.
Loop BriX statistics
In contrast to the relatively limited conformational space of regular structure elements, loop structures are much more variable. In Loop BriX, loop fragments are between 4 and 117 irregular residues long and classes are generally less populated (Figure 2B). Intriguingly, we observe a clear distinction between classes of loops connecting different secondary structure: the number of super-classes having more than 100 fragments is much lower for α-α (8) than β-β classes (20), showing less regularity for α-α classes than for β-β classes (Supplementary Figure S7). This is explained by the fact that α-helices, being cylindrical, show much more variation at their end points, while β-strands have more regular end-to-end distances.
We then examined the results of our loop classification scheme, looking at the percentage of loops we were able to classify. At the super class level our approach classified almost 90% of 6-residue loops and 45% of 14-residue loops while the success of sub-clustering in equally sized groups decreased more rapidly (Supplementary Figure S8A). We found that the sub-classification was successful up to fragments of length 16, after which no regular loop patterns could be identified (Supplementary Figure S8B).
Applications of the BriX database
The first version of the BriX database already inspired many applications in the fields of structural biology and protein design. Baeten et al. showed that proteins from the widely used Park & Levitt set could be reconstructed using BriX fragments to a global 0.48 Å RMSD accuracy, improving existing results using more limited structural alphabets (22).
Demon et al. used BriX database fragments in combination with the FoldX protein design algorithm to construct a model of murine caspase 3 and 7 in complex with substrate peptides. These models were subsequently used to explain experimentally observed differences in substrate specificity between caspase 3 and 7 (28,29).
In other recent work, we have shown that the structural space of protein–peptide interactions can be approximated using fragments from the BriX database (30). The interfaces of over 300 protein–peptide complexes from the PepX database (31) were reconstructed to within 1 Å RMSD, using observed fragment interactions to reconstruct the binding modes. The sheer size of the database allowed us to extract structural knowledge on protein–peptide interactions.
Until now, all of these services have been limited to internal use of the database. With the updated version of the BriX and Loop BriX databases, the website and the addition of the covering and bridging algorithms (see below), we open up the possibilities to use the BriX database to the scientific community at large.
DATABASE ACCESS
User interface
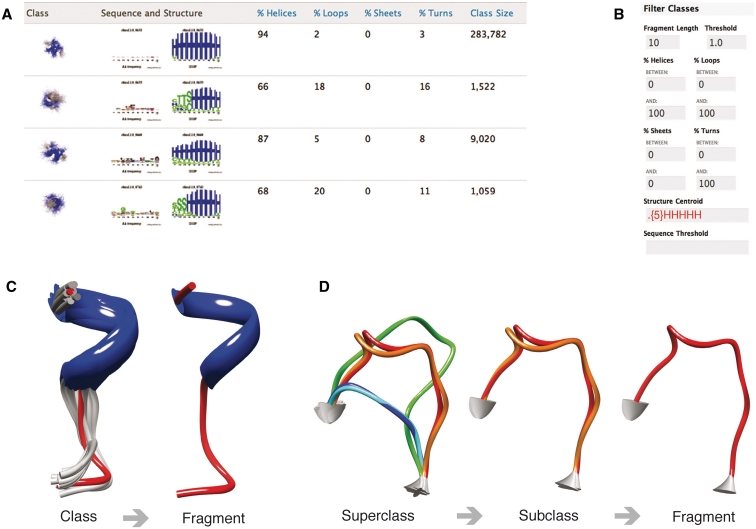
A user-friendly browsing interface is available on the website (http://brix.crg.es, Figure 3A). BriX contains two levels: the class level and the fragment level (Figure 3C). Classes can be sorted and filtered on (i) class size, (ii) fragment length (from 4 to 14 residues), (iii) clustering threshold describing the compactness of the classes, (iv) minimum and maximum percentage of helix, loop, sheet and turn content and (v) regular expressions of the amino acid sequence and secondary structure as determined by DSSP (32) (Figure 3B). For each BriX class, we generated images of the superposed fragments using Chimera (33) and logos of the sequence and structure distributions using Weblogo (34). Subsequently, the fragments of each class can be filtered on PDB ID (35), sequence or secondary structure.
Figure 3.
The BriX website (http://brix.crg.es) (A) An overview of the class level with secondary structure content and sequence and structure logos per entry. (B) A panel on the class level where a user can filter on length, threshold, sequence and secondary structure content. Similar panels are implemented at every level of the class hierarchy. (C) BriX contains two levels: the class level and the fragment level. (D) Loop BriX contains three levels: the superclass, the subclass and the fragment level.
Loop BriX contains three levels: (i) the superclass level with fragments of similar end-to-end distance and matching end residues, (ii) the subclass level with fragments of similar backbone patterns and length and finally, (iii) the fragment level (Figure 3D). The Loop BriX superclasses and subclasses can be queried with the same parameters as the BriX database plus end-to-end distance.
Query the database by covering or bridging protein structures
To explore the vast size of our database we provide two algorithms to query BriX and Loop BriX with a user-submitted structure: ‘covering’ and ‘bridging’. The covering algorithm covers backbone coordinates of the input structure with similar BriX classes. The bridging algorithm spans the distance between any pair of anchoring residues regardless of backbone coordinates in between them. This is extremely useful to derive plausible loop conformations where backbone coordinates are not present or poorly defined.
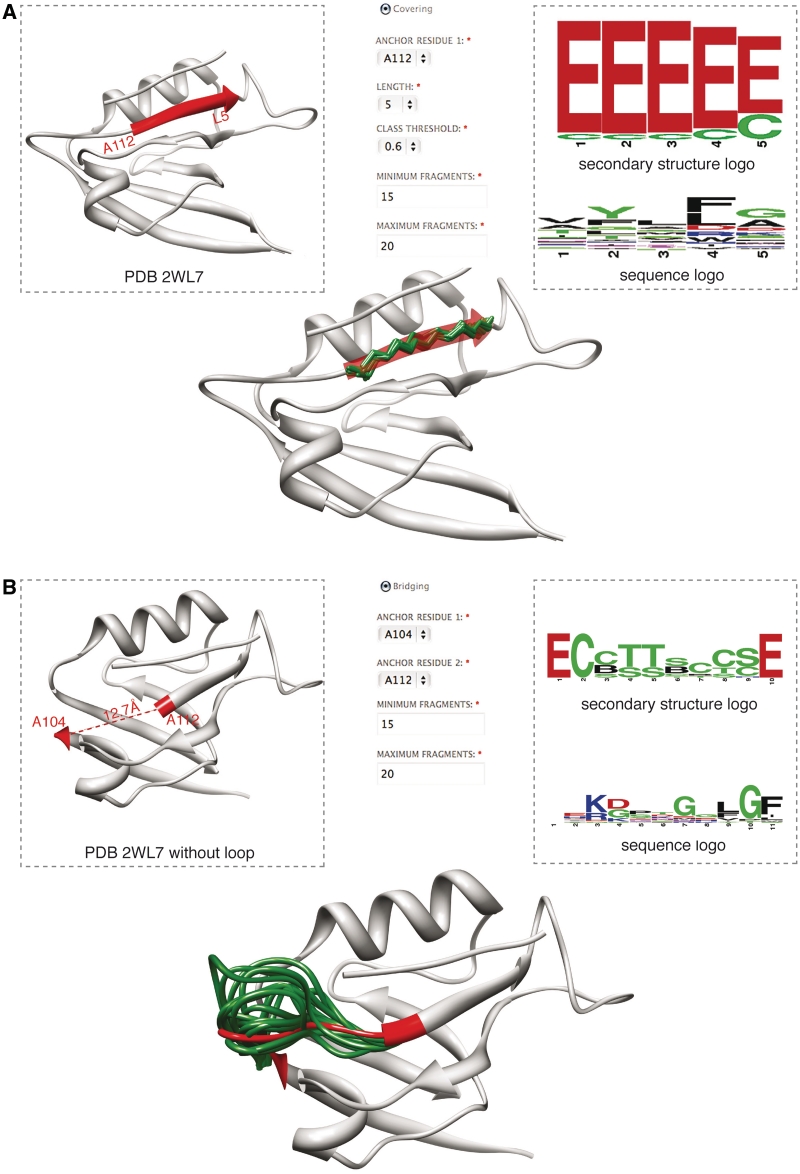
In Figure 4A, we show the application of the covering algorithm to a PDZ domain (PDB ID 2WL7), covering a part of the β-strand with classes from the BriX database. Residues 112–116 are selected for covering. The algorithm matches the selected region to the BriX classes by calculating the distance to each class centroid. Here, the user can select the class threshold that defines their compactness (0.6 Å in this example). Fragments are returned for every class having a centroid close enough to the query fragment. The user can also select the maximum number of fragments per class, the total minimum and maximum number of fragments (between 1 and 1000) and superposition thresholds are adapted accordingly. In the case of the β-strand of the PDZ, over 3000 fragments superposing with 0.6 Å are matched, of which 1000 are returned to the user as a set of downloadable fragment PDB files. Moreover, the service provides a snapshot of these fragments superposed on the query PDB as well as logos depicting sequence and structure propensities of the matched fragments, useful to derive sequence or structure relationships. Finally, the set of matching classes and fragments can be further inspected online using the previously described search interface.
Figure 4.
BriX applications: ‘covering’ and ‘bridging’. (A) Covering: an input PDZ structure (PDB: 2WL7) is shown for which the algorithm finds matching structural fragments for the β-strand starting at residue 112 in chain A (red). The algorithm returns a set of protein fragment structures (green) superposed on the β-strand, together with structure and sequence logos. (B) Bridging: the same PDB structure (PDB: 2WL7), now with a missing loop. The algorithm finds loop fragments that match the regular anchor residues 104 and 112 spanning the loop with the same end-to-end distance (green).
The bridging algorithm works in a similar fashion. To illustrate this, we removed a loop of the same PDZ domain from the input structure (Figure 4B), which is involved in binding the peptide ligand of this domain. This loop is anchored by residue 104 on the left and residue 112 on the right, spanning a gap of 12.7Å end-to-end distance. The algorithm reconstructs a backbone with fragments from the Loop BriX database between the two anchor residues. As one might expect, the results contain loops from other PDZ domains (e.g. PDB ID 1WIF), but also loops derived from proteins with unrelated SCOP classes.
Given the vastness of our database, calculations can be demanding. We allocated a dedicated cluster (40 nodes) that runs the algorithms independent from the web server.
Database availability and automated database interaction through web-based API
The BriX and Loop BriX databases are accessible through a web portal at http://brix.crg.es. The portal is built on the open-source Drupal Content Management System for full flexibility. The entire database with annotations is available for download in the SQL format, describing the relations between classes and fragments. As an additional service for automated high-throughput querying, all information contained within the BriX and Loop BriX database can be downloaded as CSV (comma-separated values) lists. For example, prompting the URL http://brix.crg.es/classes?Length=10&Structure=HHHHHHHHHH returns a CSV file containing BriX classes of length 10 with an α-helical structure. Finally, BriX will be updated automatically when new versions of the ASTRAL sets will become available.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
PhD scholarship from the Institute for Science and Innovation Flanders (IWT) (to P.V. and L.B.); Long-term exchange fellowship from the Research Foundation Flanders (FWO) (to P.V.); PhD scholarship from the EU grant Penelope FW6 (to E.V.). Funding from the EU grants 3D repertoire and the Spanish grant Centrosome3D. Funding for open access charge: EU grants 3D repertoire and the Spanish grant Centrosome3D.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors thank Almer M. van der Sloot and Joke Reumers for critical reading of the manuscript.
REFERENCES
- 1.Levitt M. Nature of the protein universe. Proc. Natl Acad. Sci. USA. 2009;106:11079–11084. doi: 10.1073/pnas.0905029106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Kopp J, Bordoli L, Battey JND, Kiefer F, Schwede T. Assessment of CASP7 predictions for template-based modeling targets. Proteins. 2007;69(Suppl. 8):38–56. doi: 10.1002/prot.21753. [DOI] [PubMed] [Google Scholar]
- 3.Murzin AG, Brenner SE, Hubbard T, Chothia C. SCOP: a structural classification of proteins database for the investigation of sequences and structures. J. Mol. Biol. 1995;247:536–540. doi: 10.1006/jmbi.1995.0159. [DOI] [PubMed] [Google Scholar]
- 4.Orengo CA, Michie AD, Jones S, Jones DT, Swindells MB, Thornton JM. CATH – a hierarchic classification of protein domain structures. Structure. 1997;5:1093–1108. doi: 10.1016/s0969-2126(97)00260-8. [DOI] [PubMed] [Google Scholar]
- 5.Fitzkee NC, Fleming PJ, Gong H, Panasik N, Street TO, Rose GD. Are proteins made from a limited parts list? Trends Biochem. Sci. 2005;30:73–80. doi: 10.1016/j.tibs.2004.12.005. [DOI] [PubMed] [Google Scholar]
- 6.Budowski-Tal I, Nov Y, Kolodny R. FragBag, an accurate representation of protein structure, retrieves structural neighbors from the entire PDB quickly and accurately. Proc. Natl Acad. Sci. USA. 2010;107:3481–3486. doi: 10.1073/pnas.0914097107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Le Q, Pollastri G, Koehl P. Structural alphabets for protein structure classification: a comparison study. J. Mol. Biol. 2009;387:431–450. doi: 10.1016/j.jmb.2008.12.044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ananthalakshmi P, Kumar CK, Jeyasimhan M, Sumathi K, Sekar K. Fragment Finder: a web-based software to identify similar three-dimensional structural motif. Nucleic Acids Res. 2005;33:W85–W88. doi: 10.1093/nar/gki353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Berkholz DS, Krenesky PB, Davidson JR, Karplus PA. Protein Geometry Database: a flexible engine to explore backbone conformations and their relationships to covalent geometry. Nucleic Acids Res. 2010;38:D320–D325. doi: 10.1093/nar/gkp1013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Samson AO, Levitt M. Protein segment finder: an online search engine for segment motifs in the PDB. Nucleic Acids Res. 2009;37:D224–D228. doi: 10.1093/nar/gkn833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Kolodny R, Koehl P, Guibas L, Levitt M. Small libraries of protein fragments model native protein structures accurately. J. Mol. Biol. 2002;323:297–307. doi: 10.1016/s0022-2836(02)00942-7. [DOI] [PubMed] [Google Scholar]
- 12.Kolodny R, Levitt M. Protein decoy assembly using short fragments under geometric constraints. Biopolymers. 2003;68:278–285. doi: 10.1002/bip.10262. [DOI] [PubMed] [Google Scholar]
- 13.Bystroff C, Shao Y. Fully automated ab initio protein structure prediction using I-SITES, HMMSTR and ROSETTA. Bioinformatics. 2002;18(Suppl. 1):S54–S61. doi: 10.1093/bioinformatics/18.suppl_1.s54. [DOI] [PubMed] [Google Scholar]
- 14.Kifer I, Nussinov R, Wolfson HJ. Constructing templates for protein structure prediction by simulation of protein folding pathways. Proteins. 2008;73:380–394. doi: 10.1002/prot.22073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bornot A, Etchebest C, de Brevern AG. A new prediction strategy for long local protein structures using an original description. Proteins. 2009;76:570–587. doi: 10.1002/prot.22370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Choi Y, Deane CM. FREAD revisited: accurate loop structure prediction using a database search algorithm. Proteins. 2010;78:1431–1440. doi: 10.1002/prot.22658. [DOI] [PubMed] [Google Scholar]
- 17.Fernandez-Fuentes N, Zhai J, Fiser A. ArchPRED: a template based loop structure prediction server. Nucleic Acids Res. 2006;34:W173–W176. doi: 10.1093/nar/gkl113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian B, Raman S, Das R, Bradley P, McCoy AJ, Read RJ, Baker D. High-resolution structure prediction and the crystallographic phase problem. Nature. 2007;450:259–264. doi: 10.1038/nature06249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Simons KT, Kooperberg C, Huang E, Baker D. Assembly of protein tertiary structures from fragments with similar local sequences using simulated annealing and Bayesian scoring functions. J. Mol. Biol. 1997;268:209–225. doi: 10.1006/jmbi.1997.0959. [DOI] [PubMed] [Google Scholar]
- 20.Pandini A, Fornili A, Kleinjung J. Structural alphabets derived from attractors in conformational space. BMC Bioinformatics. 2010;11:97. doi: 10.1186/1471-2105-11-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fitzkee NC, Fleming PJ, Rose GD. The Protein Coil Library: a structural database of nonhelix, nonstrand fragments derived from the PDB. Proteins. 2005;58:852–854. doi: 10.1002/prot.20394. [DOI] [PubMed] [Google Scholar]
- 22.Baeten L, Reumers J, Tur V, Stricher F, Lenaerts T, Serrano L, Rousseau F, Schymkowitz J. Reconstruction of protein backbones from the BriX collection of canonical protein fragments. PLoS Comput. Biol. 2008;4:e1000083. doi: 10.1371/journal.pcbi.1000083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chandonia J-M, Hon G, Walker NS, Lo Conte L, Koehl P, Levitt M, Brenner SE. The ASTRAL Compendium in 2004. Nucleic Acids Res. 2004;32:D189–D192. doi: 10.1093/nar/gkh034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Vriend G. WHAT IF: a molecular modeling and drug design program. J. Mol. Graphics. 1990;8:52–56, 29. doi: 10.1016/0263-7855(90)80070-v. [DOI] [PubMed] [Google Scholar]
- 25.Burke DF, Deane CM. Improved protein loop prediction from sequence alone. Protein Eng. 2001;14:473–478. doi: 10.1093/protein/14.7.473. [DOI] [PubMed] [Google Scholar]
- 26.Donate LE, Rufino SD, Canard LH, Blundell TL. Conformational analysis and clustering of short and medium size loops connecting regular secondary structures: a database for modeling and prediction. Protein Sci. 1996;5:2600–2616. doi: 10.1002/pro.5560051223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Espadaler J, Fernandez-Fuentes N, Hermoso A, Querol E, Aviles FX, Sternberg MJE, Oliva B. ArchDB: automated protein loop classification as a tool for structural genomics. Nucleic Acids Res. 2004;32:D185–188. doi: 10.1093/nar/gkh002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Schymkowitz J, Borg J, Stricher F, Nys R, Rousseau F, Serrano L. The FoldX web server: an online force field. Nucleic Acids Res. 2005;33:W382–W388. doi: 10.1093/nar/gki387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Demon D, Van Damme P, Vanden Berghe T, Deceuninck A, Van Durme J, Verspurten J, Helsens K, Impens F, Wejda M, Schymkowitz J, et al. Proteome-wide substrate analysis indicates substrate exclusion as a mechanism to generate caspase-7 versus caspase-3 specificity. Mol. Cell. Proteomics. 2009;8:2700–2714. doi: 10.1074/mcp.M900310-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vanhee P, Stricher F, Baeten L, Verschueren E, Lenaerts T, Serrano L, Rousseau F, Schymkowitz J. Protein-peptide interactions adopt the same structural motifs as monomeric protein folds. Structure. 2009;17:1128–1136. doi: 10.1016/j.str.2009.06.013. [DOI] [PubMed] [Google Scholar]
- 31.Vanhee P, Reumers J, Stricher F, Baeten L, Serrano L, Schymkowitz J, Rousseau F. PepX: a structural database of non-redundant protein-peptide complexes. Nucleic Acids Res. 2010;38:D545–D551. doi: 10.1093/nar/gkp893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kabsch W, Sander C. Dictionary of protein secondary structure: pattern recognition of hydrogen-bonded and geometrical features. Biopolymers. 1983;22:2577–2637. doi: 10.1002/bip.360221211. [DOI] [PubMed] [Google Scholar]
- 33.Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera – a visualization system for exploratory research and analysis. J. Comput. Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 34.Crooks GE, Hon G, Chandonia J-M, Brenner SE. WebLogo: a sequence logo generator. Genome Res. 2004;14:1188–1190. doi: 10.1101/gr.849004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Berman HM, Westbrook J, Feng Z, Gilliland G, Bhat TN, Weissig H, Shindyalov IN, Bourne PE. The Protein Data Bank. Nucleic Acids Res. 2000;28:235–242. doi: 10.1093/nar/28.1.235. [DOI] [PMC free article] [PubMed] [Google Scholar]