Abstract
The Protein Data Bank in Europe (PDBe; pdbe.org) is actively involved in managing the international archive of biomacromolecular structure data as one of the partners in the Worldwide Protein Data Bank (wwPDB; wwpdb.org). PDBe also develops new tools to make structural data more widely and more easily available to the biomedical community. PDBe has developed a browser to access and analyze the structural archive using classification systems that are familiar to chemists and biologists. The PDBe web pages that describe individual PDB entries have been enhanced through the introduction of plain-English summary pages and iconic representations of the contents of an entry (PDBprints). In addition, the information available for structures determined by means of NMR spectroscopy has been expanded. Finally, the entire web site has been redesigned to make it substantially easier to use for expert and novice users alike. PDBe works closely with other teams at the European Bioinformatics Institute (EBI) and in the international scientific community to develop new resources with value-added information. The SIFTS initiative is an example of such a collaboration—it provides extensive mapping data between proteins whose structures are available from the PDB and a host of other biomedical databases. SIFTS is widely used by major bioinformatics resources.
INTRODUCTION
The Protein Data Bank in Europe (PDBe; pdbe.org) (1) is the European partner in the Worldwide Protein Data Bank (wwPDB; wwpdb.org) (2), the international partnership that manages the Protein Data Bank (PDB) (3,4) archive of experimentally determined biomacromolecular structures. The other wwPDB partners are the Research Collaboratory for Structural Bioinformatics (RCSB) (5) and the BioMagResBank (BMRB) (6) in the USA, as well as the Protein Data Bank Japan (PDBj) (7). The four partners provide data deposition and annotation facilities for the experimental structural-biology community. This collaboration has resulted in a single, uniform archive for macromolecular structure data and has led to substantial improvements in the quality, consistency and integrity of the archive. The PDB is updated weekly with new and revised entries and is made available by all the wwPDB sites simultaneously at 0:00 UTC (Coordinated Universal Time) on Wednesdays. The archive is freely downloadable and is mirrored by many third-party sites.
The wwPDB partners each offer different and competing services to deliver the basic archival data along with value-added information, thus providing alternative and in some cases complementary ways for the user community to obtain biomacromolecular structure information. Historically, PDBe has provided the structural biology community with advanced tools and services for biomacromolecular structure search and analysis (8). PDBe has also been at the forefront of developing resources for X-ray crystallography, Nuclear Magnetic Resonance (NMR) spectroscopy and cryo-Electron Microscopy (EM). The Electron Microscopy Data Bank (EMDB; EMDataBank.org) (9,10) was established at the EBI in 2002 and is now managed and developed in collaboration with the RCSB and Baylor College of Medicine (EMDataBank.org this issue).
As the PDB approaches its 40th anniversary in 2011, PDBe has turned its attention to a fundamental problem facing the structural biology community: ‘how to make the wealth of structural data available to the larger biomedical community?’ Addressing this issue will require rethinking the ways in which structure data is delivered to users. Issues of data quality and validation are crucial to ensure that users with relatively little structural biology background can assess the quality of the data they want to use. In this article, we discuss the first steps towards providing better access to biomacromolecular structure data for expert and novice users alike. In addition, we describe new resources for value-added NMR data and the SIFTS project (11), which is the authoritative source of up-to-date residue-level annotation of protein structures in the PDB with data available in UniProt (12) and several other major biomedical databases.
PDBeXplore: A BROWSER FOR STRUCTURAL KNOWLEDGE
As a first step towards making biomacromolecular structure data available to the biomedical community, PDBe has developed an interface that allows access to the 3D-structure data based on classification systems that are familiar and intuitive to molecular biologists, biochemists and other life scientists. This marks a shift from the traditional way of accessing PDB data based on PDB accession code or by searches using information regarding, for instance, a publication, a molecule name, a sequence or a related 3D structure.
The new browsing capability can be used not only to list and sort the relevant PDB entries, but also to analyze the structural knowledge embodied in the PDB in the context of the biological knowledge represented in various biological classification systems. One of the oldest and most widely used biochemical classification systems is the ‘Enzyme Classification’ (EC) (13), which classifies the known enzymes into functional families. Based on information from the IntEnz (14) database we have developed a new interactive interface to browse PDB data in the context of the Enzyme Classification.
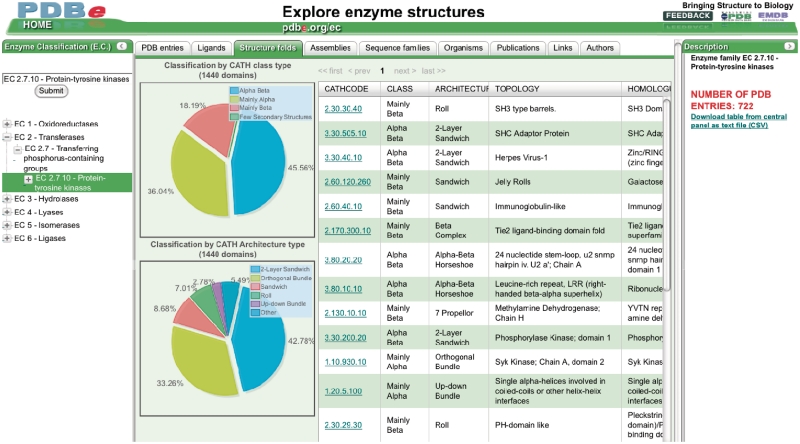
The EC browser (pdbe.org/ec) enables users to retrieve and analyze information on any or all of the enzyme structures available in the PDB. Figure 1 shows the EC-browser interface, which consists of three components: a panel that enables users to select enzymes or enzyme classes of interest; a panel that displays information about the class of enzymes selected by the user; and a central panel that presents different views on the structural information available in the PDB and related resources about the selected enzyme class, organized in a number of tabs. Each tab provides a different view on the data:
PDB entries: the default view in the browser displays information about the PDB entries (if any) that contain structures for one or more members of the selected enzyme class. The information is in a table that summarizes the most relevant aspects of each PDB entry (e.g., PDB code, EC class, experimental method, deposition date, resolution, organism name). The table can be sorted on any of these attributes by clicking the corresponding column header.
Ligands: this view displays information (e.g. ligand identifier, name and formula) about the ligand molecules found in the PDB entries for the selected enzyme class. By default, the ligands are sorted by the total number of occurrences in the PDB entries of the selected enzyme class.
Structure folds: this view displays information about the fold families [based on the CATH classification (15)] encountered in PDB entries that contain a member of the selected enzyme class. The tab also shows the distribution of the most frequent CATH classes and architectures for the selected enzyme class as pie charts.
Assemblies: this view provides information about the possible quaternary structure(s) of the selected PDB entries calculated with the program PISA (16). A tally of monomeric, homomeric and heteromeric assemblies is presented in a table. Two pie charts show a further breakdown of the composition of homomeric and heteromeric structures. This view also shows the possible quaternary structure(s) for the entries, together with (for non-monomeric structures) the accessible surface area of the complex, its buried surface area and the estimated free-energy gain upon formation of the complex.
Sequence families: this table displays information about all the Pfam (17) sequence families encountered in the PDB entries containing members of the selected enzyme class.
Organisms: the source organisms of the proteins in the selected PDB entries are listed. Two pie charts show the distribution of the most common organisms based on superkingdom (bacteria, archaea, etc.) and genus (homo, rattus, bacillus, etc.).
Publications: shows detailed information on the (primary) publications of the selected PDB entries.
Links: this view contains information from the CAS (http://www.cas.org/), GO (18), PROSITE (19) and UniProt (12) databases for the selected enzyme class.
Authors: lists the names of all the authors of the structures of the selected enzyme class in the PDB, sorted by the number of PDB entries of which they are authors. This view allows a user to quickly identify structural biologists who have (had) an interest in the selected EC class (e.g. as potential collaborators or referees).
Figure 1.
The Enzyme Classification-based PDB browser (pdbe.org/ec; see text for details).
All the data displayed in the browser is retrieved from the PDBe database in real time and all graphs are generated on the fly. Hence, the information shown is always up-to-date. The interface also allows users to download data presented in the central panel for further analysis or reporting. Browsing the structural archive in this fashion gives both expert and non-expert users an intuitive means for accessing and analyzing the wealth of information available in the PDB, using familiar biological or (bio-)chemical terms and classifications.
In addition to the EC browser, PDBe has developed two other browser modules that are based on the sequence-based protein-family classification system Pfam (17) (pdbe.org/pfam) and the structure-fold-based protein-family classification system CATH (15) (pdbe.org/cath), respectively, and further modules are under development. There is also a browser-like interface for analyzing the results of FASTA-based (20) sequence searches of the PDB (21) (pdbe.org/fasta). The functionality and interface of these browsers is very similar to that of the EC browser.
PDBprints: AT-A-GLANCE SUMMARIES OF PDB ENTRIES
In order to convey key information about a PDB entry, PDBe has designed a set of intuitive icons called PDBlogos. In addition, PDBe has introduced PDBprints (pdbe.org/pdbprints), which are sets of seven icons that convey specific bits of information in a well-defined order. On the PDBe Atlas pages, PDBprints are used to give an at-a-glance overview of the contents of an entry. PDBprints also allow for easy comparison of PDB entries listed in the result lists of the PDBe search system. In the first release of PDBprints (summer 2010), the following categories of information are included:
Primary citation: has the PDB entry been published in the literature?
Taxonomy: what is the source organism of the biomacromolecule(s) in the entry?
Sample-production technique: how was the sample of the biomacromolecule(s) obtained?
Structure-determination method: which experimental technique(s) was used to determine the structure and is the experimental data available from the PDB?
Protein content: does the entry contain any protein molecules?
Nucleic acid content: does the entry contain any nucleic acid molecules (DNA, RNA or a hybrid)?
Heterogen content: does the entry contain any ligands (such as inhibitors, co-factors, ions, metals, etc.)?
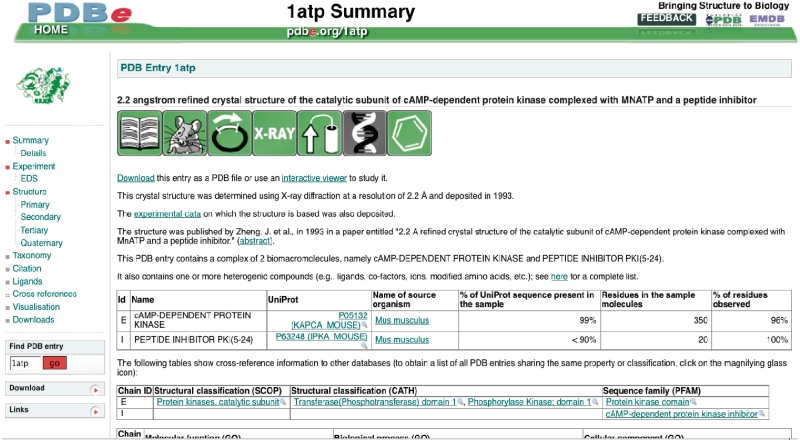
Some of the icons may have either a grey or a colored background. If the background is grey, this implies that the corresponding feature, data or information is absent or unavailable. For example, Figure 2 shows the PDBprints for PDB entry 1atp (22), which immediately reveals that 1atp is a published crystal structure of a heterologously expressed mouse protein in complex with a ligand, for which the experimental data is available. The grey icon indicates that the entry contains no nucleic acid molecules.
Figure 2.
PDBprints for PDB entry 1atp (pdbe.org/pdbprints; see text for details).
A REDESIGNED, USER-ORIENTED WEB SITE
PDBe has established itself as a provider of advanced services such as PDBeFold (23), PDBeMotif (24) and PDBePISA (16). These services are used not only by the experimental structural biology community, but also by the wider bioinformatics and biology community interested in information about biological assemblies or in comparing the folds or ligand-binding sites of structures available in the PDB. To make these services more easily accessible, we have redesigned our web site with two major objectives in mind: to make PDBe services and the PDB archive data more accessible to a broader user base, in particular novice and non-expert users, and to integrate services in a transparent manner.
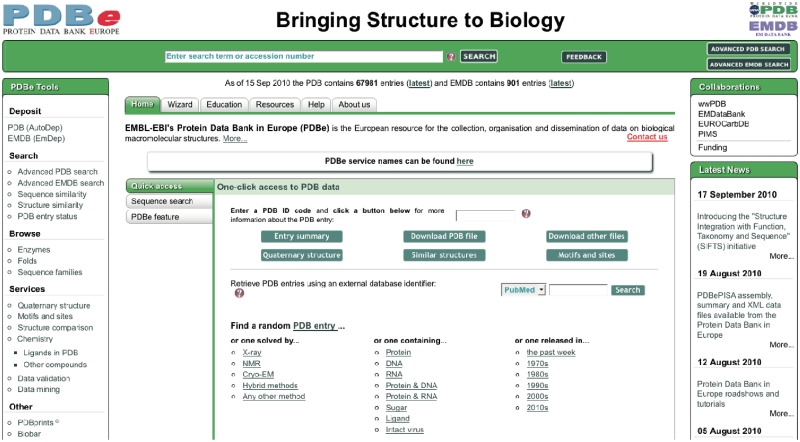
The PDBe home page (pdbe.org) was redesigned to allow easy access to the PDB data and the advanced PDBe services (Figure 3). A ‘PDBe Tools’ panel provides links to some of the most popular search and analysis tools at PDBe, organized in problem-oriented sets (‘deposit’, ‘browse’, ‘search’, etc.). The central part of the home page provides access to a wealth of information about PDBe and its resources, services and tools. By default, the page opens on the ‘Home’ tab, which offers a number of quick ways to access information for a particular PDB entry. The ‘Sequence search’ sub-tab provides easy access to the FASTA-based (21) browser interface. Users can find additional information about various PDBe resources, tools and services via the ‘PDBe feature’ sub-tab. The ‘Quick access’ sub-tab, which is displayed by default, enables users to enter a PDB code and gain single-click access to a number of commonly requested information sources about that entry. At present, these include (i) the new English-language summary Atlas page (Figure 4); (ii) the PDB-formatted file from the archive; (iii) a page with links to files related to the PDB entry [e.g. mmCIF and PDBML files of the structure, experimental data and SIFTS (11) data]; (iv) the probable quaternary structure (derived by PDBePISA); (v) similar folds in the PDB [calculated by PDBeFold (23), based on the program SSM (25)]; and (vi) analyses of sequence motifs, 3D motifs, ligand interactions, etc. from PDBeMotif (24). The ‘Quick access’ sub-tab further allows users to search for PDB entries based on an external database identifier [e.g. from PubMed (26), UniProt (12), SCOP (27), Pfam (17), CATH (15) or GO (18)]. Finally, the ‘Quick access’ sub-tab provides a number of links that give access to a random PDB entry that satisfies a criterion relating to the method used to solve the structure, the type of molecules in the entry, or when the entry was released. These links are very useful for education and outreach purposes.
Figure 3.
The redesigned PDBe home page (pdbe.org).
Figure 4.
For every PDB entry, PDBe offers a summary Atlas page (including the entry’s PDBprints) that presents vital information in simple English sentences and a few tables with cross-reference information. The other Atlas pages of an entry provide much more detailed information.
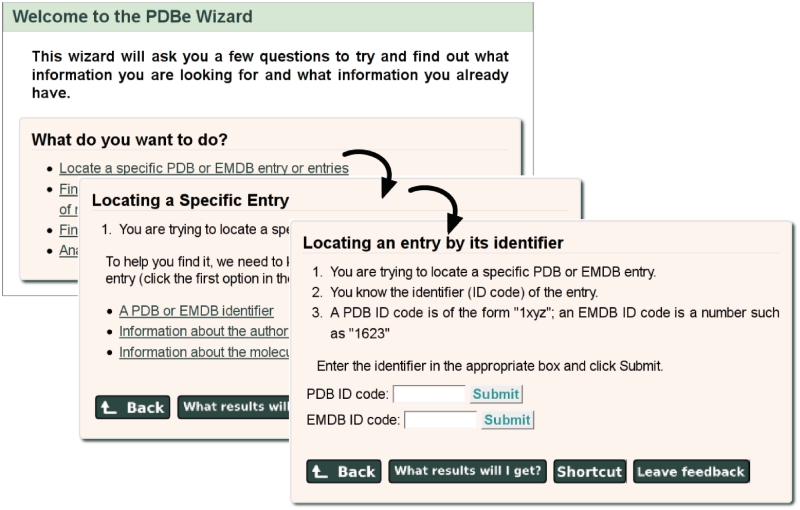
Novice users of the PDBe web site will benefit from the newly introduced Wizard. This Wizard tries to determine what a user is looking for based on answers to a series of questions. In most cases, it eventually presents users with a search form for direct access to the information or it suggests an appropriate resource or advanced service at PDBe. Users are also provided with a ‘Shortcut’ method that does not require use of the Wizard pages, should they wish to carry out similar searches in the future. Figure 5 shows an example of a series of Wizard pages.
Figure 5.
The PDBe Wizard (pdbe.org/wizard) guides users to information by asking simple questions to find out what they know and what they hope to find. The figure shows three subsequent Wizard panels that help users locate a particular PDB or EMDB entry based on its PDB code or EMDB accession number.
The search bar at the top of the PDBe home page can be used to carry out a quick search for a specific PDB or EMDB (9,10) entry, or a quick keyword search of both databases. The two databases are queried simultaneously and the search results are classified based on the categories in which the search term was present. For instance, a search term such as ‘cancer’ may occur as part of a journal title, a publication title, a molecule name, a keyword or a domain or sequence family name, etc. This facility enables users to refine their search and only find results in a category of their interest.
A final feature to help users access PDBe resources is the implementation of a number of easy-to-remember short-cut URLs (Table 1), e.g. ‘pdbe.org’ gives direct access to the PDBe home page while ‘pdbe.org/fold’ and ‘pdbe.org/pisa’ provide direct access to the PDBeFold and PDBePISA services, respectively. The URL ‘pdbe.org/1xyz’ links directly to the PDBe summary Atlas page for PDB entry ‘1xyz’, while ‘pdbe.org/download/1xyz’ gives direct access to the PDB file for that entry.
Table 1.
Shortcut URLs to some PDBe services and resources
| pdbe.org/deposit | Deposit data to the PDB using AutoDep |
| pdbe.org/1xyz | Get more information about PDB entry 1xyz |
| pdbe.org/download/1xyz | Download the PDB file of entry 1xyz |
| pdbe.org/fold | Find structures with similar folds |
| pdbe.org/pisa | Analyze assemblies, interfaces and quaternary structure |
| pdbe.org/motif | Analyze ligands and their binding properties, sequence motifs, structure motifs, etc. |
| pdbe.org/browse | Browse the structural archive |
| pdbe.org/analysis | Analyze PDB data |
| pdbe.org/pdbprints | Read about PDBprints |
| pdbe.org/nmr | Access the PDBe NMR pages |
| pdbe.org/emdb | Access the EMDB pages |
| pdbe.org/sifts | Use the SIFTS website |
| pdbe.org/wizard | Use the Wizard to find information on the PDBe website |
| pdbe.org/resources | Find out more about PDBe resources |
STRUCTURE INTEGRATION WITH FUNCTION, TAXONOMY AND SEQUENCE
The European Bioinformatics Institute (EBI) is home to a number of core bioinformatics databases and services that provide data relevant to the biomedical field. As part of the EBI, PDBe is in a unique position to enhance the annotation of biomacromolecular structures with data from other biological databases by cross-referencing and mapping to in-house resources. The ‘Structure Integration with Function, Taxonomy and Sequence’ initiative (SIFTS; pdbe.org/sifts) (11) is a close collaboration between the PDBe and UniProt (12) teams, with the goal of improving the integration of protein structure and sequence data. The project was started in 2001 and has resulted in the development of a robust mechanism for enhancing annotations and exchanging data between the major structure- and sequence-based resources.
The SIFTS procedure (S.V. et al., unpublished data) identifies the correct cross-reference in the UniProt database for every protein in a PDB entry. All the data for residue-level mapping between a PDB entry and the corresponding UniProt entry is generated using automated procedures once all the taxonomy and UniProt cross-reference information has been identified. To validate the mapping, the data is loaded into the PDBe database where data integrity checks are performed independently of the mapping process. The mapping information is enriched with cross-reference information from the NCBI taxonomy database (26,28), IntEnz (14), CATH (12), SCOP (27), InterPro (29), Pfam (15) and PubMed (26). This process is based either on information from the corresponding UniProt entry or from the links available to the PDB entry from the corresponding databases. While this process is very effective in gathering and cross-mapping these data resources, in relation to GO (18) terms it can introduce spurious mappings. This is due to the fact that all the GO terms are mapped onto the complete sequence in a UniProt entry whereas a PDB entry may only contain a fragment or domain to which the GO annotation may not be applicable. To address this problem, PDBe has developed an improved mapping process for GO terms in collaboration with the InterPro and GOA teams at the EBI. The new process uses InterProScan (30) and considers only the domains present in the PDB entry to map the corresponding GO terms (18). All cross-reference data is made freely available in tab-delimited files and XML files from the PDBe ftp area (pdbe.org/sifts/ftp). SIFTS data is used by major bioinformatics resources such as RCSB (5), PDBsum (31), Pfam (17), SCOP (27), InterPro (29), several DAS server providers (32) and many research and service groups around the world to provide cross-reference information on their web pages. Table 2 shows statistics for the PDB entries that have SIFTS-based cross-references as of September 2010.
Table 2.
SIFTS statistics of the number of PDB entries with cross-reference information (September 2010)
| Total PDB entries processed | 67 981 |
|---|---|
| Entries with no possible UniProt cross-reference | 3691 |
| Entries with UniProt cross-reference | 64 073 |
| Entries with residue-level mapping | 64 073 |
| Entries awaiting export | 217 |
| Entries with NCBI taxonomy identifier | 64 785 |
| Entries with cross-reference to InterPro | 63 292 |
| Entries with Pfam family annotation | 62 210 |
| Entries with cross-reference to Gene Ontology terms | 55 918 |
| Entries with primary citation with PubMed identifier | 54 524 |
| Entries with SCOP cross-reference | 38 078 |
| Entries with assigned CATH identifier | 35 279 |
| Entries with assigned EC classification | 28 978 |
NEW RESOURCES FOR NMR-RELATED DATA
PDBe has worked closely with the NMR community and BMRB to improve the data quality of NMR depositions in the PDB. NMR spectroscopy is an important structure-determination technique but has suffered from a lack of standards and tools, which has limited the reliability of the deposited data. Capturing experimental data is key to enhancing the reliability of NMR structures in the PDB. The deposition of restraints derived from the experimental data (such as NOE-based distant restraints) has been mandatory since February 2008, and by the end of 2010 the deposition of chemical shift information will also become mandatory.
To facilitate the deposition of NMR models, experimental data, restraints and other metadata, PDBe has developed the ‘Entry Completion Interface’ (ECI) (33). The software is based on the CCPN (34) framework and enables pooling of all NMR-related data in one project. The CCPN FormatConverter (34) can be used within ECI to import data from the output of commonly used NMR software. ECI also carries out basic validation of chemical shifts against standard values. The finalized CCPN project can be uploaded to PDBe using AutoDep (33,35). AutoDep also accepts chemical shifts as a separate file in NMR-STAR (V3.1) format and allows the input of referencing information for the relevant nuclei. In that case, an additional validation step is carried out to check the correspondence of the atom nomenclature between the coordinate and the chemical shift data. The chemical shift data is automatically forwarded to the BMRB for further annotation and archiving.
The PDBe Atlas pages contain details about the underlying experiments for every PDB entry. In the case of NMR entries, these pages have been redesigned to provide access to a wealth of publicly available information, some of which is unique to PDBe (Figure 6). For every NMR entry for which the appropriate data is available, the following information is provided (Table 3 lists the number of NMR entries for which each kind of information is available, as of September 2010):
The VASCO (36) validation report. This program may suggest a systematic correction of the deposited chemical shifts and also identifies any chemical shift outliers based on statistics that take into account the atom type, secondary structure and average accessible surface area of the atom in the ensemble. In the future, we intend to provide VASCO as a web service for validation of chemical shifts prior to deposition and at any stage of the structure-determination process.
For ensembles of protein NMR structures, the most representative conformer as identified by analysis with the program OLDERADO (37) is presented. The full OLDERADO report, which gives more detailed information about the clustering and domain organization of the ensemble, is also available (Figure 7).
RECOORD (38) is a database of recalculated NMR structure ensembles hosted at PDBe. Each RECOORD page contains a comparison of ensemble quality scores for both the originally deposited and the recalculated structures, as well as the coordinates of the recalculated ensembles.
Chemical shift data available at BMRB, linked uniquely for jointly deposited data and by sequence similarity search.
Deposited and remediated restraints for most NMR structures from the NRG-GRID (39).
Validation reports generated by the CING suite (nmr.cmbi.ru.nl/NRG-CING), containing Ramachandran statistics, identification of violated restraints, per-residue quality analysis, etc.
Reports for the NMR ensembles included in the re-refined NMR structure database DRESS (40).
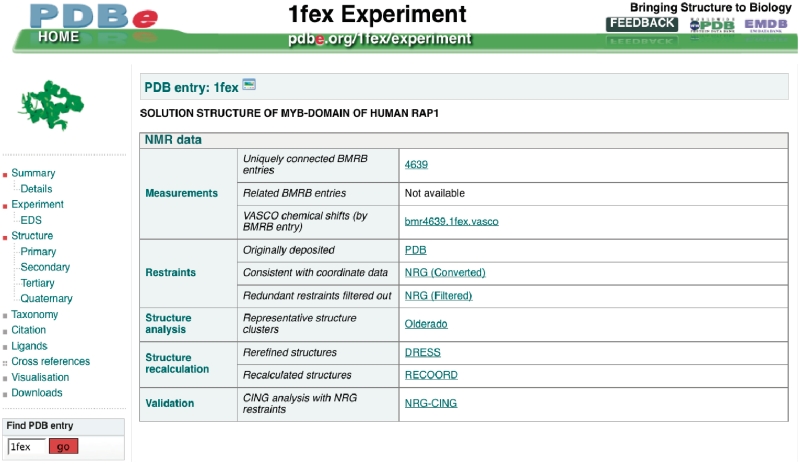
Figure 6.
One of the Atlas pages for PDB entry 1fex (42), showing NMR-related information and links.
Table 3.
Number of NMR-based PDB entries for which additional information is available through the PDBe Atlas pages (September 2010)
| NMR entries in the PDB | 8592 |
| NMR entries with OLDERADO report | 6440 |
| NMR entries with NRG restraints | 5864 |
| NMR entries with CING report | 5816 |
| NMR entries with chemical shifts available at BMRB | 4665 |
| NMR entries with VASCO report | 2303 |
| NMR entries in the RECOORD database | 539 |
| NMR entries in the DRESS resource | 99 |
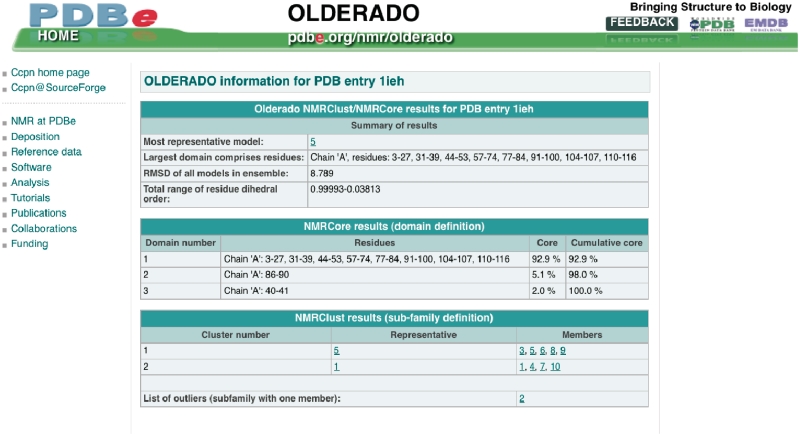
Figure 7.
Summary of OLDERADO (36) information for PDB entry 1ieh (43). The core residues used for the clustering are listed in the panel ‘NMRCore results’, and the final clustering is in the panel ‘NMRClust results’. Clicking on the links for the models will return a PDB file containing only the coordinates for that model.
FUTURE DEVELOPMENTS
PDBe works closely with its wwPDB partners, the structural biology community and bioinformatics resources at the EBI and elsewhere to improve the quality and consistency of the data in the PDB. It is actively involved in the wwPDB X-ray and NMR validation task forces and is implementing the recommendations of the X-ray validation task force (41). This validation pipeline will be used by all wwPDB partners, both to validate new depositions to the PDB and to assess the quality of existing entries. The validation data will be crucial to help experts and novices alike to access structural information that is reliable. In the next few years, PDBe will also endeavour to provide new ways to access and integrate structural and related data, especially for non-expert users. Simultaneously, PDBe will continue to develop advanced services aimed more specifically at the structural biology community.
FUNDING
PDBe gratefully acknowledges the support of the European Molecular Biology Laboratory (EMBL), the Wellcome Trust (grant number 088944), the European Union (213010 and 226073), the UK Biotechnology and Biological Sciences Research Council (BB/C512110/1, BB/G022577/1, BB/E007511/1), and the National Institutes of Health (R01GM079429-01A1). Funding for open access charge: Wellcome Trust.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors wish to thank all collaborators and partners in the EBI, EMBL, wwPDB, EMDB, CCPN, CCP4, CCDC and other collaborative efforts, as well as the structural biology community for depositing their structures and experimental data in the PDB and EMDB.
REFERENCES
- 1.Velankar S, Best C, Beuth B, Boutselakis CH, Cobley N, Sousa Da Silva AW, Dimitropoulos D, Golovin A, Hirshberg M, John M, et al. PDBe: Protein Data Bank in Europe. Nucleic Acids Res. 2010;38:D308–D317. doi: 10.1093/nar/gkp916. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berman H, Henrick K, Nakamura H, Markley JL. The worldwide Protein Data Bank (wwPDB): ensuring a single, uniform archive of PDB data. Nucleic Acids Res. 2007;35:D301–D303. doi: 10.1093/nar/gkl971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernstein FC, Koetzle TF, Williams GJ, Meyer EF, Jr, Brice MD, Rodgers JR, Kennard O, Shimanouchi T, Tasumi M. The Protein Data Bank: a computer-based archival file for macromolecular structures. J. Mol. Biol. 1977;112:535–542. doi: 10.1016/s0022-2836(77)80200-3. [DOI] [PubMed] [Google Scholar]
- 4.Berman HM. The Protein Data Bank: a historical perspective. Acta Crystallogr. 2008;A64:88–95. doi: 10.1107/S0108767307035623. [DOI] [PubMed] [Google Scholar]
- 5.Kouranov A, Xie L, de la Cruz J, Chen L, Westbrook J, Bourne PE, Berman HM. The RCSB PDB information portal for structural genomics. Nucleic Acids Res. 2006;34:D302–D305. doi: 10.1093/nar/gkj120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ulrich EL, Akutsu H, Doreleijers JF, Harano Y, Ioannidis YE, Lin J, Livny M, Mading S, Maziuk D, Miller Z, et al. BioMagResBank. Nucleic Acids Res. 2008;36:D402–D408. doi: 10.1093/nar/gkm957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Standley DM, Kinjo AR, Kinoshita K, Nakamura H. Protein structure databases with new web services for structural biology and biomedical research. Brief. Bioinform. 2008;9:276–285. doi: 10.1093/bib/bbn015. [DOI] [PubMed] [Google Scholar]
- 8.Tagari M, Tate J, Swaminathan GJ, Newman R, Naim A, Vranken W, Kapopoulou A, Hussain A, Fillon J, Henrick K, et al. E-MSD: improving data deposition and structure quality. Nucleic Acids Res. 2006;34:D287–D290. doi: 10.1093/nar/gkj163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tagari M, Newman R, Chagoyen M, Carazo J, Henrick K. New electron microscopy database and deposition system. Trends Biochem. Sci. 2002;27:589. doi: 10.1016/s0968-0004(02)02176-x. [DOI] [PubMed] [Google Scholar]
- 10.Henrick K, Newman R, Tagari M, Chagoyen M. EMDep: a web-based system for the deposition and validation of high-resolution electron microscopy macromolecular structural information. J. Struct. Biol. 2003;144:228–237. doi: 10.1016/j.jsb.2003.09.009. [DOI] [PubMed] [Google Scholar]
- 11.Velankar S, McNeil P, Mittard-Runte V, Suarez A, Barrell D, Apweiler R, Henrick K. E-MSD: an integrated data resource for bioinformatics. Nucleic Acids Res. 2005;33:D262–D265. doi: 10.1093/nar/gki058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.UniProt Consortium. The Universal Protein Resource (UniProt) 2009. Nucleic Acids Res. 2009;37:D169–D174. doi: 10.1093/nar/gkn664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Enzyme Nomenclature 1992 [A, ISBN 0-12-227164-5 (hardback), 0-12-227165-3 (paperback)]. This supplement is as close as possible to the published version [see Eur. J. Biochem., 1999, 264, 610–650] [Google Scholar]
- 14.De Matos P, Alcantara R, Dekker A, Ennis M, Hastings J, Haug K, Spiteri I, Turner S, Steinbeck C. Chemical Entities of Biological Interest: an update. Nucleic Acids Res. 2010;38:D249–D254. doi: 10.1093/nar/gkp886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Greene LH, Lewis TE, Addou S, Cuff A, Dallman T, Dibley M, Redfern O, Pearl F, Nambudiry R, Reid A, et al. The CATH domain structure database: new protocols and classification levels give a more comprehensive resource for exploring evolution. Nucleic Acids Res. 2007;35:D291–D297. doi: 10.1093/nar/gkl959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Krissinel E, Henrick K. Inference of macromolecular assemblies from crystalline state. J. Mol. Biol. 2007;372:774–797. doi: 10.1016/j.jmb.2007.05.022. [DOI] [PubMed] [Google Scholar]
- 17.Finn RD, Tate J, Mistry J, Coggill PC, Sammut SJ, Hotz HR, Ceric G, Forslund K, Eddy SR, Sonnhammer ELL, et al. The Pfam protein families database. Nucleic Acids Res. 2008;36:D281–D288. doi: 10.1093/nar/gkm960. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Barrell D, Dimmer E, Huntley R, Binns D, O’Donovan C, Apweiler R. The GOA database in 2009—an integrated Gene Ontology Annotation resource. Nucleic Acids Res. 2009;37:D396–D403. doi: 10.1093/nar/gkn803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulo N, Bairoch A, Bulliard V, Cerutti L, Cuche BA, de Castro E, Lachaize C, Langendijk-Genevaux PS, Sigrist CJ. The 20 years of PROSITE. Nucleic Acids Res. 2008;36:D245–D249. doi: 10.1093/nar/gkm977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lipman DJ, Pearson WR. Rapid and sensitive protein similarity searches. Science. 1985;227:1435–1441. doi: 10.1126/science.2983426. [DOI] [PubMed] [Google Scholar]
- 21.Bhagat J, Tanoh F, Nzuobontane E, Laurent T, Orlowski J, Roos M, Wolstencroft K, Aleksejevs S, Stevens R, et al. BioCatalogue: a universal catalogue of web services for the life sciences. Nucleic Acids Res. 2010;38:W689–W694. doi: 10.1093/nar/gkq394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng J, Trafny EA, Knighton DR, Xuong NH, Taylor SS, Ten Eyck LF, Sowadski JM. 2.2 Å refined crystal structure of the catalytic subunit of cAMP-dependent protein kinase complexed with MnATP and a peptide inhibitor. Acta Crystallogr. 1993;D49:362–365. doi: 10.1107/S0907444993000423. [DOI] [PubMed] [Google Scholar]
- 23.Krissinel E, Henrick K. Secondary-structure matching (SSM), a new tool for fast protein structure alignment in three dimensions. Acta Crystallogr. 2004;D60:2256–2268. doi: 10.1107/S0907444904026460. [DOI] [PubMed] [Google Scholar]
- 24.Golovin A, Henrick K. MSDmotif: exploring protein sites and motifs. BMC Bioinformatics. 2008;9:312. doi: 10.1186/1471-2105-9-312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Krissinel E, Henrick K. Common subgraph isomorphism detection by backtracking search. Software: Practice and Experience. 2004;34:591–607. [Google Scholar]
- 26.Sayers EW, Barrett T, Benson DA, Bryant SH, Canese K, Chetvernin V, Church DM, DiCuccio M, Edgar R, Federhen S, et al. Database resources of the National Center for Biotechnology Information. Nucleic Acids Res. 2009;37:D5–D15. doi: 10.1093/nar/gkn741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Andreeva A, Howorth D, Chandonia JM, Brenner SE, Hubbard TJ, Chothia C, Murzin AG. Data growth and its impact on the SCOP database: new developments. Nucleic Acids Res. 2008;36:D419–D425. doi: 10.1093/nar/gkm993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2009;37:D26–D31. doi: 10.1093/nar/gkn723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hunter S, Apweiler R, Attwood TK, Bairoch A, Bateman A, Binns D, Bork P, Das D, Daugherty L, Duquenne L, et al. InterPro: the integrative protein signature database. Nucleic Acids Res. 2009;37:D211–D215. doi: 10.1093/nar/gkn785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Quevillon E, Silventoinen V, Pillai S, Harte N, Mulder N, Apweiler R, Lopez R. InterProScan: protein domains identifier. Nucleic Acids Res. 2005;33:W116–W120. doi: 10.1093/nar/gki442. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Laskowski RA. PDBsum new things. Nucleic Acids Res. 2009;37:D355–D359. doi: 10.1093/nar/gkn860. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Dowell RD, Jokerst RM, Day A, Eddy SR, Stein L. The distributed annotation system. BMC Bioinformatics. 2001;2:7. doi: 10.1186/1471-2105-2-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Penkett CJ, van Ginkel G, Velankar S, Swaminathan J, Ulrich EL, Mading S, Stevens TJ, Fogh RH, Gutmanas A, Kleywegt GJ, et al. Straightforward and complete deposition of NMR data to the PDBe. J. Biomol. NMR. 2010;48:85–92. doi: 10.1007/s10858-010-9439-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Vranken WF, Boucher W, Stevens TJ, Fogh RH, Pajon A, Llinas M, Ulrich EL, Markley JL, Ionides J, Laue ED. The CCPN data model for NMR spectroscopy: development of a software pipeline. Proteins. 2005;59:687–696. doi: 10.1002/prot.20449. [DOI] [PubMed] [Google Scholar]
- 35.Tagari M, Tate J, Swaminathan GJ, Newman R, Naim A, Vranken WF, Kapopoulou A, Hussain A, Fillon J, Henrick K, et al. E-MSD: improving data deposition and structure quality. Nucleic Acids Res. 2006;34:D287–D290. doi: 10.1093/nar/gkj163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Rieping W, Vranken WF. Validation of archived chemical shifts through atomic coordinates. Proteins. 2010;78:2482–2489. doi: 10.1002/prot.22756. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kelley LA, Sutcliffe MJ. OLDERADO: On Line Database of Ensemble Representatives And DOmains. Protein Sci. 1997;6:2628–2630. doi: 10.1002/pro.5560061215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nederveen AJ, Doreleijers JF, Vranken W, Miller Z, Spronk CAEM, Nabuurs SB, Guntert P, Livny M, Markley JL, Nilges M, et al. RECOORD: A recalculated coordinate database of 500+ proteins from the PDB using restraints from the BioMagResBank. Proteins. 2005;59:662–672. doi: 10.1002/prot.20408. [DOI] [PubMed] [Google Scholar]
- 39.Doreleijers JF, Vranken WF, Schulte C, Lin J, Wedell JR, Penkett CJ, Vuister GW, Vriend G, Markley JL, Ulrich EL. The NMR restraints grid at BMRB for 5,266 protein and nucleic acid PDB entries. J. Biomol. NMR. 2009;45:389–396. doi: 10.1007/s10858-009-9378-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Nabuurs SB, Nederveen AJ, Vranken W, Doreleijers JF, Bonvin AMJJ, Vuister GW, Vriend G, Spronk CAEM. DRESS: a Database of REfined Solution NMR Structures. Proteins. 2004;55:483–486. doi: 10.1002/prot.20118. [DOI] [PubMed] [Google Scholar]
- 41.Berman H, Kleywegt GJ, Nakamura H, Markley J, Burley SK. Safeguarding the integrity of protein archive. Nature. 2010;463:425. doi: 10.1038/463425c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hanaoka S, Nagadoi A, Yoshimura S, Aimoto S, Li B, de Lange T, Nishimura Y. NMR structure of the hRap1 Myb motif reveals a canonical three-helix bundle lacking the positive surface charge typical of Myb DNA-binding domains. J. Mol. Biol. 2001;312:167–175. doi: 10.1006/jmbi.2001.4924. [DOI] [PubMed] [Google Scholar]
- 43.Mayer KL, Stone MJ. NMR solution structure and receptor peptide binding of the CC chemokine eotaxin-2. Biochemistry. 2000;39:8382–8395. doi: 10.1021/bi000523j. [DOI] [PubMed] [Google Scholar]