Abstract
Autophagy is a process of self-digestion generally observed in eukaryotes and has been shown to play crucial roles for survival under starvation and removal of deleterious substances. Despite great advances that have been made, many problems in mechanisms of autophagy remain unsolved. As a large number of autophagy-related proteins are identified in each species, a database that collects data, identifies their homologs in other species and makes them available will contribute to research advancement. As no such resources exist, we built the Autophagy database (http://tp-apg.genes.nig.ac.jp/autophagy) to provide basics, up-to-date information on relevant literature, and a list of autophagy-related proteins and their homologs in 41 eukaryotes. From the database, the user can search for proteins by keywords or sequences to obtain a wealth of data including functional and structural information and find possible functional homologs of proteins whose functions have been demonstrated in other species. As proteins that bind the phospholipid, phosphatidyl inositol 3-phosphate (PI3P) are essential for autophagy to proceed, we carried out an original analysis to identify probable PI3P-binding proteins, and made the list available from the database. The database is expected to give impetus to further research on autophagy by providing basic and specialized data on the subject.
INTRODUCTION
Autophagy is a process of self-digestion observed in all eukaryotes examined (1), indicating its essentiality in this domain of life. Macroautophagy, heretofore autophagy, involves the formation of a membrane (autophagosome) around a region of the cytoplasm, sequestering macromolecules such as proteins and organelles, and the fusion of the resultant vesicle with a vacuole (in plants and fungi) or a lysosome (other eukaryotes) in which the contents are subsequently degraded. Through autophagy, starving cells nonselectively degrade materials within their own cells to produce amino acids that are then used for survival.
Another essential function of autophagy in higher eukaryotes is to selectively remove deleterious substances such as misfolded proteins, abnormal organelles and intracellular pathogens: defects in autophagy in animals are known to cause neurodegeneration and tumorigenesis (2). The induction of autophagy in response to starvation at least partially explains the age-lengthening effect of caloric restriction (3). Intriguingly, autophagy is regulated in coordination with apoptosis: cells either survive by self-digestion or die by self-killing. Elucidation of autophagy is thus not only of academic, but also of medical interest.
Although research on autophagy made great advances after a pioneering genetic study on Saccharomyces cerevisiae (4), a number of outstanding issues remain. For instance, how autophagosomes form is not well understood and how autophagy selectively removes certain substances has been mostly unexplored (3). A clue to the former problem was provided by the finding that PI3P generated on membrane helps to localize autophagy-related proteins (5). In addition, mitochondrial membranes were identified as a source of autophagosomal membranes in rat cells (6). Recent structural studies of autophagy-related proteins in S. cerevisiae gave insights into mechanisms of selective autophagy (7,8).
For effective utilization of available information, a specialized database will be useful. Specifically, functional and structural information on a multitude of autophagy-related proteins can be effectively utilized in studies of other species if probable functional homologs are identified and made publicly available. To our knowledge, autophagy databases covering multiple species do not exist, although an excellent database specializing in human autophagy is available (http://autophagy.lu/index.test.html). To promote research on autophagy, we built the Autophagy Database that covers 41 eukaryotes and made it publicly available with the aim to provide useful information to researchers.
EXPLANATIONS AND LITERATURE INFORMATION
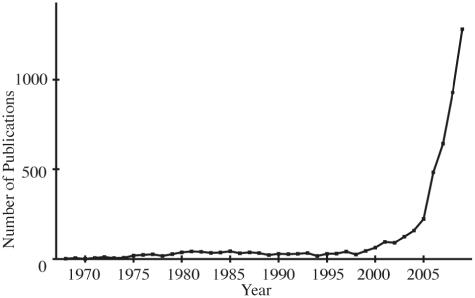
To provide an introduction to non-specialists and results of past and cutting-edge research to specialists, the database gives an overview and bibliographic information on autophagy. The top page provides a brief description of autophagy and figures of autophagic processes, while the ‘Overview section’ gives explanations on the database, statistics and a list of recent reviews on the subject. In the ‘New Refs section’, the user can get a list of recently published papers on the subject: relevant publications that appeared on a user-specified day or month are displayed. We plotted the annual breakdown of the number of papers on autophagy in the database (Figure 1). The figure shows a clear upward trend from year 2000 in the number of publications, reflecting rapid advances in autophagy-related research in recent years. As the accelerating pace makes it desirable to expeditiously perform evaluations of recent publications and make them publicly available, we devised a mechanism to allow users to evaluate each paper and show the rating. The system prompts the user to rate each article on a scale of 1–3, with the larger numbers signifying higher evaluation and displays the average score. We deliberately designed a simple system so that many users will participate in the undertaking and thereby enhance the value of the database.
Figure 1.
Annual breakdown of the number of publications on autophagy. The number of autophagy-related publications published in each year from 1968 to 2009 in the Autophagy Database is graphed.
LIST OF AUTOPHAGY-RELATED PROTEINS AND THEIR HOMOLOGS
From review articles (9,10), we obtained a list of 133 proteins in S. cerevisiae (baker’s yeast), Homo sapiens and Mus musculus (mouse), whose involvement in autophagy have been experimentally demonstrated. Consultation with another paper (11) yielded 499 orthologs of autophagy-related proteins. Using these ‘reviewed’ and ‘orthologous’ proteins as queries, we identified homologs in 41 entirely sequenced eukaryotes, including a number of model organisms such as baker’s yeast, Drosophila, H. sapiens and M. musculus (Table 1). Specifically, we ran PSI-BLAST (12) with three iterations setting the E-value cut-off at 0.1, and from the final results selected 1531 homologs that have the E-value ≤1e-100. We thus have a total of 2163 proteins that are either known to be involved in autophagy or are homologous to autophagy-related proteins.
Table 1.
Eukaryotes covered by the database
| Classification | Species (common name) |
|---|---|
| Excavata | Trypanosoma brucei |
| Fungi | Ashbya gossypii |
| Aspergillus fumigatus | |
| Aspergillus nidulans | |
| Aspergillus oryzae | |
| Candida albicans | |
| Candida glabrata | |
| Chaetomium globosum | |
| Coccidioides immitis | |
| Cryptococcus neoformans var. neoformans | |
| Debaryomyces hansenii | |
| Gibberella zeae | |
| Kluyveromyces lactis | |
| Magnaporthe grisea | |
| Neurospora crassa | |
| Saccharomyces cerevisiae (baker's yeast) | |
| Schizosaccharomyces pombe | |
| Ustilago maydis | |
| Yarrowia lipolytica | |
| Plantae | Arabidopsis thaliana (thale cress) |
| Oryza sativa (rice) | |
| Invertebrata | Aedes aegypti (yellow fever mosquito) |
| Anopheles gambiae (mosquito) | |
| Apis mellifera (honey bee) | |
| Caenorhabditis elegans (nematode) | |
| Drosophila melanogaster | |
| Chordata | Bos taurus (cow) |
| Canis familiaris (dog) | |
| Cavia porcellus (guinea pig) | |
| Danio rerio (zebrafish) | |
| Felis catus (cat) | |
| Gallus gallus (chicken) | |
| Gasterosteus aculeatus (threespine stickleback) | |
| Homo sapiens (human) | |
| Macaca mulatta (rhesus monkey) | |
| Monodelphis domestica (opossum) | |
| Mus musculus (mouse) | |
| Ornithorhynchus anatinus (platypus) | |
| Oryctolagus cuniculus (European rabbit) | |
| Oryzias latipes (Japanese medaka) | |
| Rattus norvegicus (rat) |
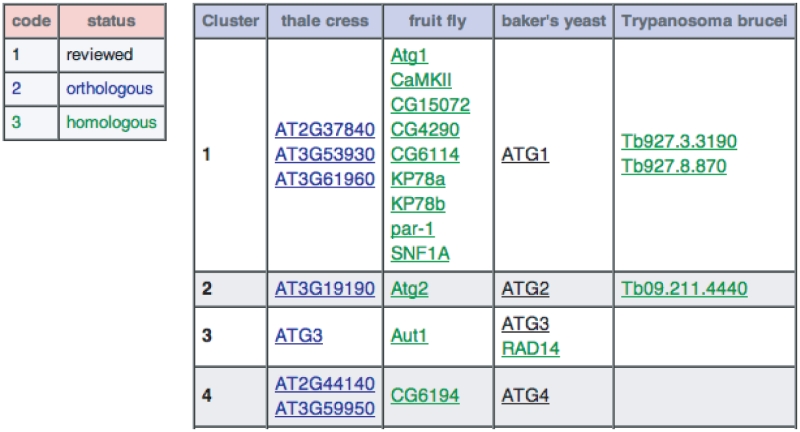
In the ‘Homologs section’, the user can select any of the 41 species and display the corresponding homologs (Figure 2). This feature enables the user to find which proteins in each species are possible functional homologs of a particular autophagy-related protein in other species. The homologs were divided into clusters according to the query proteins used. For example, the top-most two Trypanosoma brucei proteins in the figure, Tb927.3.3190 and Tb927.8.870, were identified as homologs of the S. cerevisiae protein, atg1p, and therefore were classified into the cluster of the ATG1 product, i.e. cluster #1. As homologs are displayed in green, while reviewed and orthologous proteins are shown in black and blue, respectively, the user can easily select the ones for further examination. This interspecies correspondence of autophagy-related proteins is a unique feature of the Autophagy Database.
Figure 2.
Result of a homolog search. A portion of the result of a ‘Homologs’ search in four eukaryotes is displayed. The left-most column of the main table represents the cluster number, while the remaining four columns show homologs in the four specified species. Protein symbols in black, blue and green are ‘reviewed’ proteins, orthologous proteins and other homologs identified by PSI-BLAST, respectively.
The list of proteins of each species can be viewed in the ‘Protein List section’. The user can get the cluster number, synonyms, the gene name, the Entrez gene ID, the GI number of National Center for Biotechnology Information (NCBI), the NCBI Protein Accession number together with the version, description and the PDB IDs. Clicking the symbol column opens a new window displaying detailed functions, UniProt accession number(s), UniGene ID of NCBI, related paper(s) and protein sequence as well as other information. Gene ID, GI, Protein Accession number, PDB IDs are also clickable and are linked to the relevant data.
SEARCH ENGINES FOR PROTEINS
The database provides rapid keyword and homology search engines for autophagy-related proteins. Inputting keywords such as ‘cvt yeast’ in Keyword Search results in a display of proteins possessing the keywords. If the user has a particular protein in mind, e.g. Atg19, use of the exact match option limits the search and produces fewer hits. Alternatively, the user can input an amino acid sequence in the ‘Homology Search section’ to find a protein. The rapidity and precision of searches are an intrinsic advantage a specialized database has over general ones: searches in general-purpose databases often take time and produce massive hits, from which the user must laboriously select the desired data, while searches in a more limited set are faster and tend to give rise to less false positives.
ORIGINAL RESEARCH
Besides giving free, quick access to up-to-date knowledge, the Autophagy Database aims to promote autophagy research by carrying out original analyses and making the results available. As a starter, we identified proteins that are likely to bind the phospholipid, PI3P. We carried out this analysis because autophagosome formation requires PI3P enriched in a compartment that is dynamically connected to the endoplasmic reticulum (ER), but what PI3P-binding proteins are involved and in what way remain to be elucidated (5).
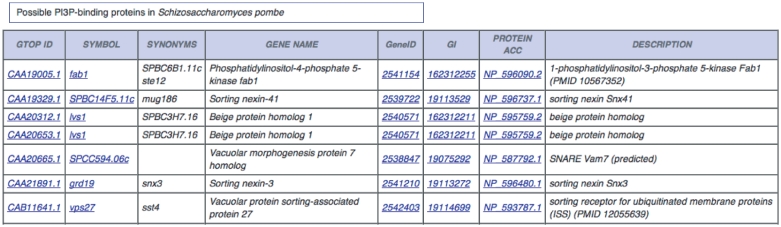
We searched for proteins with the FYVE domain (13) and considered all of them to be PI3P-binding proteins. By contrast, proteins with the PX domain may bind phospholipids other than PI3P (14). We selected a set of 35 proteins with the PX domain that are known to specifically bind PI3P (‘positive set’) and another set of nine PX-domain containing proteins that show specific binding to other phospholipids (‘negative set’). (We listed the primary accession numbers of the proteins in the sets in the ‘Help section’.) We surmised that the sequence proximity a PX domain-containing protein to either set would accurately predict the phospholipid binding specificity. A jackknife test revealed that the BLAST identification of the closest homolog in the two sets predicts which PX-domain containing proteins specifically bind PI3P at 93% precision. We thus selected proteins with the PX domain and predicted them to specifically bind PI3P if their sequences are more similar to proteins in the positive set than those in the negative set. The set of proteins that probably bind PI3P consists of proteins with the PX domain that satisfy the sequence proximity criterion and those with the FYVE domain. The list of proteins in a user-specified species that probably bind PI3P is displayed (Figure 3).
Figure 3.
Possible PI3P-binding proteins. The first seven in the list of possible PI3P-binding proteins in S. pombe together with user-specified categories of information obtained in the ‘Original Analyses section’ are presented.
FUTURE DIRECTIONS
Recent research taking a proteomic approach has vastly expanded the repertoire of autophagy-related proteins. For instance, 409 human proteins were reported to be possibly interacting with 32 autophagy-related proteins (15). We consider it desirable to identify the probable orthologs of newly found autophagy-related proteins in other eukaryotes and made them publicly available. Doing so will enable researchers to experimentally investigate the actual functions of the listed proteins.
Structural studies on autophagy-related proteins are revealing molecular mechanisms of autophagy (7,8). Many proteins consist not only of structural segments, but also of intrinsically disordered (ID) segments. Although ID segments are those that do not assume unique three-dimensional structures by themselves, many of them are functionally important. To facilitate structural determination as well as identifying functionally important ID segments, it is desirable to precisely identify the ID and structural segments. The recently developed DICHOT system divides the entire protein sequences into the two classes with precision in excess of 97% (16). We plan to apply the DICHOT system to autophagy-related proteins, accurately classify them into ID and structural segments and made the results available. A preliminary application of the DICHOT system revealed that the preApe1-binding domain of Atg19 in S. cerevisiae (7) is an ID segment, suggesting that ID segments in autophagy-related proteins may be involved in important protein-to-protein interactions. Structural segments identified by the DICHOT system contain novel (‘cryptic’) structural segments, i.e. those with no significant sequence homology with those of PDB structures. Such cryptic structural segments will be obvious targets of future structural studies. The public availability of clear structural classification of autophagy-related proteins is expected to energize research.
FUNDING
Funding for open access charge: The Targeted Proteins Research Program (TPRP) from the Ministry of Education, Culture, Sports, Science and Technology (MEXT), Japan.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
We would like to express our gratitude to many researchers whose comments and other help were instrumental in building the database, especially Drs. Y. Ohsumi, K. Suzuki, H. Nakatogawa, N. Mizushima, T. Yoshimori, T. Noda, Y. Kamada and I. Tanida.
REFERENCES
- 1.Klionsky DJ, Emr SD. Autophagy as a regulated pathway of cellular degradation. Science. 2000;290:1717–1721. doi: 10.1126/science.290.5497.1717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mizushima N. Autophagy: process and function. Genes Dev. 2007;21:2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 3.Mizushima N, Levine B, Cuervo AM, Klionsky DJ. Autophagy fights disease through cellular self-digestion. Nature. 2008;451:1069–1075. doi: 10.1038/nature06639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tsukada M, Ohsumi Y. Isolation and characterization of autophagy-defective mutants of Saccharomyces cerevisiae. FEBS Lett. 1993;333:169–174. doi: 10.1016/0014-5793(93)80398-e. [DOI] [PubMed] [Google Scholar]
- 5.Axe EL, Walker SA, Manifava M, Chandra P, Roderick HL, Habermann A, Griffiths G, Ktistakis NT. Autophagosome formation from membrane compartments enriched in phosphatidylinositol 3-phosphate and dynamically connected to the endoplasmic reticulum. J. Cell. Biol. 2008;182:685–701. doi: 10.1083/jcb.200803137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hailey DW, Rambold AS, Satpute-Krishnan P, Mitra K, Sougrat R, Kim PK, Lippincott-Schwartz J. Mitochondria supply membranes for autophagosome biogenesis during starvation. Cell. 2010;141:656–667. doi: 10.1016/j.cell.2010.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Watanabe Y, Noda NN, Kumeta H, Suzuki K, Ohsumi Y, Inagaki F. Selective transport of alpha-mannosidase by autophagic pathways: structural basis for cargo recognition by Atg19 and Atg34. J. Biol. Chem. 2010;285:30026–30033. doi: 10.1074/jbc.M110.143545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yamaguchi M, Noda NN, Nakatogawa H, Kumeta H, Ohsumi Y, Inagaki F. Autophagy-related protein 8 (Atg8) family interacting motif in Atg3 mediates the Atg3-Atg8 interaction and is crucial for the cytoplasm-to-vacuole targeting pathway. J. Biol. Chem. 2010;285:29599–29607. doi: 10.1074/jbc.M110.113670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hara T, Mizushima N. [Regulators of mammalian cellular autophagy] Tanpakushitsu Kakusan Koso. 2006;51:1484–1489. [PubMed] [Google Scholar]
- 10.Ohsumi Y. [Autophagy related genes in yeast, S. cerevisiae] Tanpakushitsu Kakusan Koso. 2006;51:1453–1456. [PubMed] [Google Scholar]
- 11.Meijer WH, van der Klei IJ, Veenhuis M, Kiel JA. ATG genes involved in non-selective autophagy are conserved from yeast to man, but the selective Cvt and pexophagy pathways also require organism-specific genes. Autophagy. 2007;3:106–116. doi: 10.4161/auto.3595. [DOI] [PubMed] [Google Scholar]
- 12.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res. 1997;25:3389–3402. doi: 10.1093/nar/25.17.3389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Gaullier JM, Simonsen A, D'Arrigo A, Bremnes B, Stenmark H, Aasland R. FYVE fingers bind PtdIns(3)P. Nature. 1998;394:432–433. doi: 10.1038/28767. [DOI] [PubMed] [Google Scholar]
- 14.Ellson CD, Andrews S, Stephens LR, Hawkins PT. The PX domain: a new phosphoinositide-binding module. J. Cell. Sci. 2002;115:1099–1105. doi: 10.1242/jcs.115.6.1099. [DOI] [PubMed] [Google Scholar]
- 15.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010;466:68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fukuchi S, Homma K, Minezaki Y, Gojobori T, Nishikawa K. Development of an accurate classification system of proteins into structured and unstructured regions that uncovers novel structural domains: its application to human transcription factors. BMC Struct. Biol. 2009;9:26. doi: 10.1186/1472-6807-9-26. [DOI] [PMC free article] [PubMed] [Google Scholar]