Abstract
It is 12 years since the IMGT/HLA database was first released, providing the HLA community with a searchable repository of highly curated HLA sequences. The HLA complex is located within the 6p21.3 region of human chromosome 6 and contains more than 220 genes of diverse function. Many of the genes encode proteins of the immune system and are highly polymorphic. The naming of these HLA genes and alleles and their quality control is the responsibility of the WHO Nomenclature Committee for Factors of the HLA System. Through the work of the HLA Informatics Group and in collaboration with the European Bioinformatics Institute, we are able to provide public access to this data through the web site http://www.ebi.ac.uk/imgt/hla/. Regular updates to the web site ensure that new and confirmatory sequences are dispersed to the HLA community, and the wider research and clinical communities.
INTRODUCTION
The IMGT/HLA database was established to provide a locus specific database (LSDB) for the allelic sequences of the genes in the HLA system, also known as the human Major Histocompatibility Complex (MHC). This complex of >4 Mb is located within the 6p21.3 region of the short arm of human chromosome 6 and contains in excess of 220 genes (1). The core genes of interest in the HLA system are 21 highly polymorphic HLA genes, whose protein products mediate the host response to infectious disease and influence the outcome of cell and organ transplants. With a nomenclature spanning over 50 genes and currently over 5000 alleles, there is an obvious need for a LSDB to curate these highly polymorphic variants. The sequencing of HLA alleles began in the late 1970′s predominantly using protein-based techniques to determine the sequences of HLA class I allotypes. The first complete HLA class I allotype sequence, B7.2, now known as B*07:02:01, was published in 1979 (2). The first HLA class II allele defined by DNA sequencing, DRA*01:01, followed in 1982 (3). The first HLA DNA sequences or alleles were named by the WHO Nomenclature Committee for Factors of the HLA System (4) in 1987. At that time 12 class I alleles and nine class II alleles were named: in the first 8 months of 2010 the Nomenclature Committee was able to assign names to 1165 alleles.
The dissemination of new allele names and sequences is of paramount importance in the clinical setting. The first public release of the IMGT/HLA database was made on the 16th December 1998 (5). Since then the database has been updated every 3 months, in a total of 51 releases, to include all the publicly available sequences officially named by the WHO Nomenclature Committee at the time of release.
The database was first available as the HLA Sequence Databank (HLA-DB) (6), which allowed the periodic publication of HLA class I (7–10) and class II (11–16) sequence alignments in a variety of journals. By 1995, the first distribution of the HLA sequence alignments was made online through the web pages of the Tissue Antigen Laboratory at the Imperial Cancer Research Fund (ICRF), London, UK. This work transferred to the Anthony Nolan Research Institute (ANRI) in 1996 where it continues to this day as part of the IMGT/HLA database and the hla.alleles.org web site.
IMGT/HLA data sources
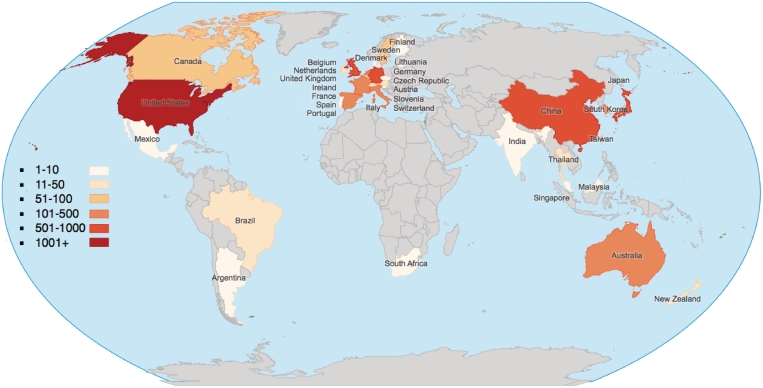
The IMGT/HLA database receives submissions from laboratories across the world (Figure 1). These submissions are curated and analyzed, and if they meet the strict requirements an official allele designation is assigned. The IMGT/HLA database is the official repository for the WHO Nomenclature Committee for factors of the HLA System, and is the only way of receiving an official allele designation for a sequence. The sequence is then incorporated into the next 3-monthly release of the database. Since its release in December 1998 the database has received nearly 9000 submissions, from around 600 submitters (Figure 1). These submissions come from a variety of sources; the majority are from routine HLA Typing laboratories or commercial organizations performing contract HLA typing for large haematopoietic stem cell donor registries. Further data has been submitted following large-scale genome sequencing projects. All submissions must meet strict acceptance criteria before the sequence receives an official designation; ∼3% of the submissions received fail to meet these criteria and are rejected. In addition, all the submissions received by the IMGT/HLA database are also available from the EMBL-Bank/GenBank/DDBJ collaboration (17–19). The EMBL-Bank entries also contain database cross-references to the IMGT/HLA entries.
Figure 1.
World map showing the source and volume of IMGT/HLA submissions by country.
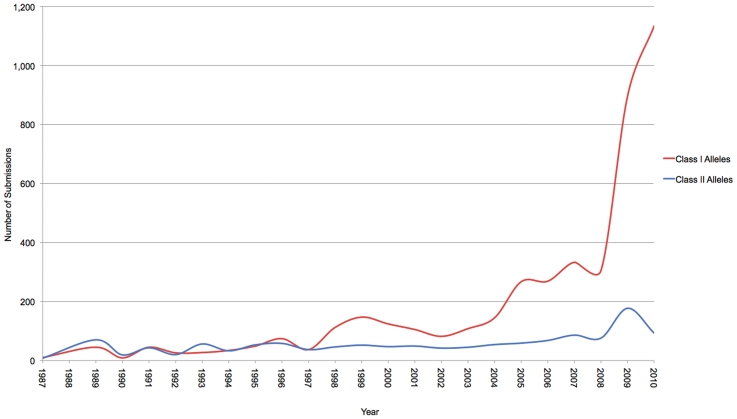
The past few years have seen a dramatic increase in the numbers of submissions seen and processed, with the number of novel allele sequences identified each year rising rapidly from around 300 in 2008 to over 1000 in 2009. This trend looks set to continue, with over 1200 novel alleles being reported in the first 9 months of 2010 (Figure 2). This is because of the increased affordability and availability of the sequencing-based typing (SBT) technology as the method of choice for HLA typing, with the consequence of this high-resolution typing being the determination of many novel HLA sequences. A notable increase in volume has been from sequences originating from China. Prior to 2008, the database only had 28 submitters located in China; we now have over 70 submitters. The volume of submissions has also increased. Up to 2008, we averaged only 18 submissions a year from China, we are now averaging nearly 200 a year, a 10-fold increase.
Figure 2.
Graph of the number of submissions to the IMGT/HLA database by year. The recent surge in the number of submissions received by the database is clearly shown. The values listed for 2010 are up to the end of September 2010, and do not represent a full year.
Another change in the data source has been the type of submission received. In the early days of the database, we received very few full-length or genomic sequences, now with improved sequencing techniques we are getting a much larger number of both full length and genomic sequences covering a range of genes. These submissions cover both new and confirmatory sequences, and the database welcomes both. Confirmatory sequences are important as they verify the existence of the single nucleotide polymorphisms (SNPs) found in many novel alleles. The confirmatory sequences often extend the sequence of an allele beyond that currently held in the database, where many alleles sequences only cover the minimum length required. Over the last 2 years just <40% of the submissions to the database have been confirmatory sequences.
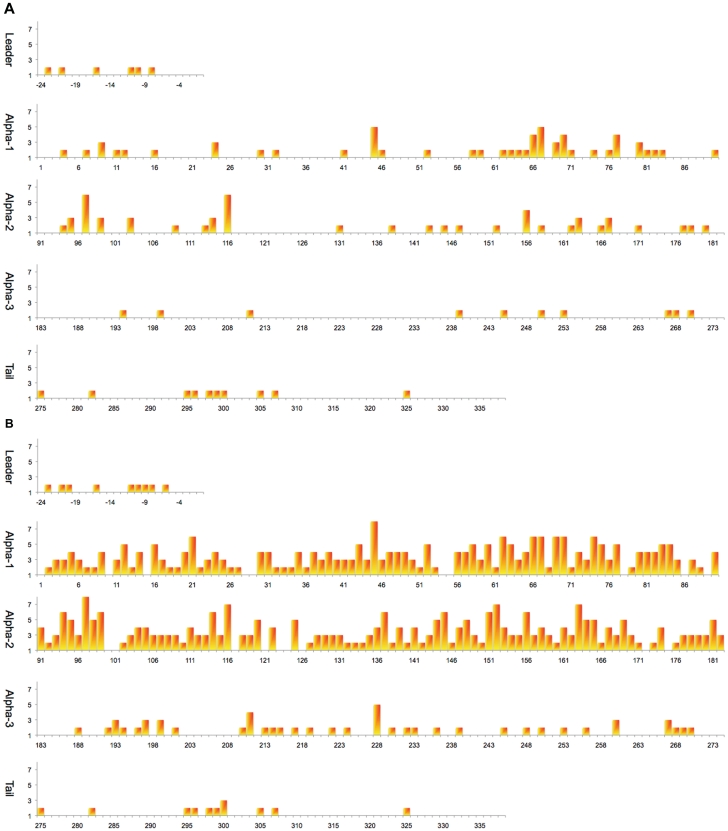
The increase in the number of submissions has also seen a change in the type of new alleles seen. Over 97% of new alleles now being submitted are derived from SNPs. In contrast, in 2000, ∼20% of new alleles identified were based on motif shuffling. This is most likely due to the methods used to identify alleles at this time that were largely based on sequence-specific oligonucleotide probes (20). Nowadays sequencing-based typing methods are used extensively to perform HLA typing and this allows for the easy identification of novel SNPs (Figure 3).
Figure 3.
Heat maps of the polymorphic amino acid positions in HLA-B. The two sets of maps show the increase in the number of polymorphic positions identified between the first release of the database in 1998 (A) and the latest release in 2010 (B). The x-axis is the amino acid position and the y-axis the number of different amino acids seen at that position.
New HLA nomenclature
In April 2010, the official nomenclature used to name HLA alleles was changed (21). The nomenclature changes were needed, as the existing system could no longer cope with the number of allele variants found in some allele families. The convention of using a four-digit code to distinguish HLA alleles that differed in the proteins they encoded was introduced in the 1987 HLA Nomenclature Report (4). Since that time additional digits have been added, and prior to the change, an allele name could be composed of four, six or eight digits dependent on its sequence. Each pair of digits was used to describe the allele, the first two digits described the allele family, which often corresponds to the serological antigen carried by the allotype. The third and fourth digits were assigned in the order in which the sequences had been determined. Alleles whose numbers differed in the first four digits differed by one or more nucleotide substitutions that changed the amino-acid sequence of the encoded protein. Alleles that differed only by synonymous nucleotide substitutions within the coding sequence were distinguished by the use of the fifth and sixth digits. Alleles that only differed by sequence polymorphisms in introns or in the 5′- and 3′-untranslated regions that flanked the exons and introns were distinguished by the use of the seventh and eighth digits. To deal with the ever increasing number of HLA alleles described it was decided to introduce colons (:) into the allele names to act as delimiters of the separate fields.
For some users the changes to the nomenclature were minor, to others like HLA Typing Laboratories and Donor Registries, this change in nomenclature had a major impact on their informatics systems. The IMGT/HLA database helped to co-ordinate the move to the new nomenclature by providing conversion lists and tools to help identify alleles in both the new and old nomenclature. The nomenclature officially changed on the 1 April 2010. To aid our users in preparing for this change, the database provided conversion tables for 9 months prior to the release. These tables allowed users to see what the changes would be and how they would impact on their own systems. The database also provided online tools for the conversion of allele names, as well as links to external software designed for the conversion of large data sets from the old to new nomenclature (22).
Further information on HLA nomenclature can be found at the IMGT/HLA database’s sister site http://hla.alleles/org. This site concentrates on HLA nomenclature, whereas the IMGT/HLA database is more focussed on sequence data. There is some overlap between the sites, but with a different prime focus each site can deliver a different set of data and downloadable content that may not be suitable for the other.
Tools available at IMGT/HLA
The IMGT/HLA database provides a large number of tools for the analysis of HLA sequences. These tools are either custom written for the database or are incorporated into existing tools on the EBI web site (23,24).
Sequence alignments—access to alignment tool, which filters pre-generated alignments to the users’ specification. Provides alignments at the protein, cDNA and gDNA level.
Allele queries—access to detailed information on any HLA Allele, including information on the ethnic origin of the source, database cross-references and seminal publications. This information is also available through integration with EBI’s SRS search engine (25).
Sequence search tools—integration into EBI’s suite of search tools including FASTA (26) and BLAST (27).
Downloads—access to a FTP directory containing all the data from the current and previous releases in a variety of commonly used formats like FASTA, MSF and PIR.
Cell Queries—a detailed a searchable database of all the source material characterized in the submissions.
Primer search tools—a simple search tool allowing users to update primer hit pattern tables against each release of the database.
Ambiguous allele combinations—the use of SBT as a method for defining the HLA type is well documented, most SBT typing strategies currently employed use the exons 2 and 3 sequences for HLA class I analysis and exon 2 alone for HLA class II analysis. Due to the heterozygous nature of the SBT analysis the combinations of many pairs of alleles may give an ambiguous typing result. The document includes a list of all alleles that are identical over exons 2 + 3 for HLA class I and exon 2 for HLA class II.
FUTURE DEVELOPMENTS
The challenge for the database is to keep up with the continuing increase in sequence information, develop new tools for the visualization of the sequences whilst maintaining the high standards set in the presentation and quality of the HLA sequences and nomenclature to the research community. The database aims to continually develop new tools and refine existing tools to meet this challenge. Some of our planned future developments include heat maps of polymorphic positions and a tool for the graphical comparison of two allele sequences, to highlight how changes to the DNA sequences affect the protein structure and binding to proteins.
CONCLUSIONS
The IMGT/HLA database provides a centralized resource for everybody interested, clinically or scientifically, in the HLA system. The database and accompanying tools allow the study of all HLA alleles from a single site on the World Wide Web. It should aid in the management and continual expansion of HLA nomenclature, providing an ongoing resource for the WHO Nomenclature Committee. The earliest version of the IMGT/HLA database, December 1998, included only 964 alleles, covering 24 genes and was limited to much simpler tools and interfaces. The latest release, July 2010, contained over 5300 alleles for 34 genes, with this number set to grow as the database continues to receive and name over a thousand new alleles each year. The expansion of the database content has been reflected in its use, in 1999 the web site averaged just over 1500 visitors per month; in 2010 this had increased to over 20 000 visitors viewing over 50 000 pages per month. The challenge for the database is to keep up with this increase in sequences, develop new tools for the visualization of the sequences whilst maintaining the high standards set in the presentation and quality of the HLA sequences and nomenclature to the research community.
LICENSING
The IMGT/HLA database is covered by the Creative Commons Attribution-NoDerivs Licence, which is applicable to all copyrightable parts of the database, which includes the sequence alignments. This means that users are free to copy, distribute, display and make commercial use of the databases in all legislations, provided they give the appropriate credit (28,29). If users intend to distribute a modified version of the data in any form, then they must ask us for permission; this can be done by contacting hla@alleles.org for further details of how modified data can be reproduced.
FUNDING
Histogenetics; Abbott Molecular Laboratories Inc.; Bio-Rad; Gen-Probe, Invitrogen by Life Technologies; European Federation for Immunogenetics; Innogenetics; One Lambda Inc.; Olersup SSP; American Society for Histocompatibility and Immunogenetics; Anthony Nolan; BAG Healthcare; Be the Match Foundation; Innogenetics; the Marrow Foundation; the National Marrow Donor Program; Rose and Zentrum Knochenmarkspender-Register Deutschland. Imperial Cancer Research Fund, (now Cancer Research UK to IMGT/HLA database project); EU Biotech (grant BIO4CT960037 to IMGT/HLA database project). Funding for open access charge: Anthony Nolan, a charitable organisation.
Conflict of interest statement. None declared.
ACKNOWLEDGEMENTS
The authors would like to thank Angie Dahl of the Be The Match Foundation, for her work in securing ongoing funding for the database. They would like to thank all of the individuals and organizations that support our work financially.
APPENDIX - ACCESS AND CONTACT
IMGT/HLA Homepage: http://www.ebi.ac.uk/imgt/hla/
IMGT/HLA FTP Site: ftp://ftp.ebi.ac.uk/pub/databases/imgt/mhc/hla/
Contact: hla@alleles.org
REFERENCES
- 1.Horton R, Wilming L, Rand V, Lovering RC, Bruford EA, Khodiyar VK, Lush MJ, Povey S, Talbot CC, Jr, Wright MW, et al. Gene map of the extended human MHC. Nat. Rev. Genet. 2004;5:889–899. doi: 10.1038/nrg1489. [DOI] [PubMed] [Google Scholar]
- 2.Orr HT, Lopez de Castro JA, Lancet D, Strominger JL. Complete amino acid sequence of a papain-solubilized human histocompatibility antigen, HLA-B7. 2. Sequence determination and search for homologies. Biochemistry. 1979;18:5711–5720. doi: 10.1021/bi00592a030. [DOI] [PubMed] [Google Scholar]
- 3.Lee JS, Trowsdale J, Travers PJ, Carey J, Grosveld F, Jenkins J, Bodmer WF. Sequence of an HLA-DR alpha-chain cDNA clone and intron-exon organization of the corresponding gene. Nature. 1982;299:750–752. doi: 10.1038/299750a0. [DOI] [PubMed] [Google Scholar]
- 4.Bodmer WF, Albert E, Bodmer JG, Dupont B, Mach B, Mayr WR, Sasazuki T, Schreuder GMT, Svejgaard A, Terasaki PI. Nomenclature for factors of the HLA system, 1987. In: Dupont B, editor. Immunobiology of HLA. Vol. 1. New York: Springer; 1989. pp. 72–79. [Google Scholar]
- 5.Robinson J, Bodmer JG, Malik A, Marsh SGE. Development of the international immunogenetics HLA database. Human Immunology. 1998;59:17. [Google Scholar]
- 6.Marsh SGE, Bodmer JG. HLA Class II Sequence Databank. Human Immunology. 1993;36:44. [Google Scholar]
- 7.Zemmour J, Parham P. HLA class I nucleotide sequences, 1991. Tissue Antigens. 1991;37:174–180. doi: 10.1111/j.1399-0039.1991.tb01869.x. [DOI] [PubMed] [Google Scholar]
- 8.Zemmour J, Parham P. HLA class I nucleotide sequences, 1992. Tissue Antigens. 1992;40:221–228. doi: 10.1111/j.1399-0039.1992.tb02049.x. [DOI] [PubMed] [Google Scholar]
- 9.Arnett KL, Parham P. HLA class I nucleotide sequences, 1995. Tissue Antigens. 1995;46:217–257. doi: 10.1111/j.1399-0039.1995.tb03124.x. [DOI] [PubMed] [Google Scholar]
- 10.Mason PM, Parham P. HLA class I region sequences, 1998. Tissue Antigens. 1998;51:417–466. doi: 10.1111/j.1399-0039.1998.tb02983.x. [DOI] [PubMed] [Google Scholar]
- 11.Marsh SGE, Bodmer JG. HLA-DRB nucleotide sequences, 1990. Immunogenetics. 1990;31:141–144. doi: 10.1007/BF00211548. [DOI] [PubMed] [Google Scholar]
- 12.Marsh SGE, Bodmer JG. HLA class II nucleotide sequences, 1991. Tissue Antigens. 1991;37:181–189. doi: 10.1111/j.1399-0039.1991.tb01870.x. [DOI] [PubMed] [Google Scholar]
- 13.Marsh SGE, Bodmer JG. HLA class II nucleotide sequences, 1992. Tissue Antigens. 1992;40:229–243. doi: 10.1111/j.1399-0039.1992.tb02050.x. [DOI] [PubMed] [Google Scholar]
- 14.Marsh SGE, Bodmer JG. HLA class II region nucleotide sequences, 1994. Eur. J. Immunogenet. 1994;21:519–551. doi: 10.1111/j.1744-313x.1994.tb00223.x. [DOI] [PubMed] [Google Scholar]
- 15.Marsh SGE, Bodmer JG. HLA class II region nucleotide sequences, 1995. Tissue Antigens. 1995;46:258–280. doi: 10.1111/j.1399-0039.1995.tb03125.x. [DOI] [PubMed] [Google Scholar]
- 16.Marsh SGE. HLA class II region sequences, 1998. Tissue Antigens. 1998;51:467–507. doi: 10.1111/j.1399-0039.1998.tb02984.x. [DOI] [PubMed] [Google Scholar]
- 17.Benson DA, Karsch-Mizrachi I, Lipman DJ, Ostell J, Sayers EW. GenBank. Nucleic Acids Res. 2010;38:D46–D51. doi: 10.1093/nar/gkp1024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kaminuma E, Mashima J, Kodama Y, Gojobori T, Ogasawara O, Okubo K, Takagi T, Nakamura Y. DDBJ launches a new archive database with analytical tools for next-generation sequence data. Nucleic Acids Res. 2010;38:D33–D38. doi: 10.1093/nar/gkp847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Leinonen R, Akhtar R, Birney E, Bonfield J, Bower L, Corbett M, Cheng Y, Demiralp F, Faruque N, Goodgame N, et al. Improvements to services at the European Nucleotide Archive. Nucleic Acids Res. 2010;38:D39–D45. doi: 10.1093/nar/gkp998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Erlich H, Bugawan T, Begovich AB, Scharf S, Griffith R, Saiki R, Higuchi R, Walsh PS. HLA-DR, DQ and DP typing using PCR amplification and immobilized probes. Eur. J. Immunogenet. 1991;18:33–55. doi: 10.1111/j.1744-313x.1991.tb00005.x. [DOI] [PubMed] [Google Scholar]
- 21.Marsh SGE, Albert ED, Bodmer WF, Bontrop RE, Dupont B, Erlich HA, Fernandez-Vina M, Geraghty DE, Holdsworth R, Hurley CK, et al. Nomenclature for factors of the HLA system, 2010. Tissue Antigens. 2010;75:291–455. doi: 10.1111/j.1399-0039.2010.01466.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Mack SJ, Hollenbach JA. Allele Name Translation Tool and Update NomenCLature: software tools for the automated translation of HLA allele names between successive nomenclatures. Tissue Antigens. 2010;75:457–461. doi: 10.1111/j.1399-0039.2010.01477.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Goujon M, McWilliam H, Li W, Valentin F, Squizzato S, Paern J, Lopez R. A new bioinformatics analysis tools framework at EMBL-EBI. Nucleic Acids Res. 2010;38(Suppl.):W695–W699. doi: 10.1093/nar/gkq313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McWilliam H, Valentin F, Goujon M, Li W, Narayanasamy M, Martin J, Miyar T, Lopez R. Web services at the European Bioinformatics Institute-2009. Nucleic Acids Res. 2009;37:W6–W10. doi: 10.1093/nar/gkp302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Etzold T, Ulyanov A, Argos P. SRS: information retrieval system for molecular biology data banks. Methods Enzymol. 1996;266:114–128. doi: 10.1016/s0076-6879(96)66010-8. [DOI] [PubMed] [Google Scholar]
- 26.Pearson WR, Lipman DJ. Improved tools for biological sequence comparison. Proc. Natl Acad. Sci. USA. 1988;85:2444–2448. doi: 10.1073/pnas.85.8.2444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Altschul SF, Gish W, Miller W, Myers EW, Lipman DJ. Basic local alignment search tool. J. Mol. Biol. 1990;215:403–410. doi: 10.1016/S0022-2836(05)80360-2. [DOI] [PubMed] [Google Scholar]
- 28.Robinson J, Malik A, Parham P, Bodmer JG, Marsh SGE. IMGT/HLA database–a sequence database for the human major histocompatibility complex. Tissue Antigens. 2000;55:280–287. doi: 10.1034/j.1399-0039.2000.550314.x. [DOI] [PubMed] [Google Scholar]
- 29.Robinson J, Waller MJ, Fail SC, McWilliam H, Lopez R, Parham P, Marsh SGE. The IMGT/HLA database. Nucleic Acids Res. 2009;37:D1013–D1017. doi: 10.1093/nar/gkn662. [DOI] [PMC free article] [PubMed] [Google Scholar]