Abstract
The NADPH-dependent protochlorophyllide (Pchlide) oxidoreductase (POR) is unique because it is a photoenzyme that requires light for its catalytic activity and uses Pchlide itself as a photoreceptor. In Arabidopsis, there are three structurally related PORs, denoted PORA, PORB, and PORC. The import of one of them, PORA, into plastids of cotyledons is substrate dependent. This substrate dependence is demonstrated in intact seedlings of wild-type Arabidopsis and two mutants, xantha2, which is devoid of Pchlide, and flu, which upon redarkening rapidly accumulates Pchlide. In true leaves, PORA uptake does not require the presence of Pchlide. The organ specificity of the substrate-dependent import of PORA reveals a means of controlling plastid protein translocation that is closely associated with a key step in plant development, the light-dependent transformation of cotyledons from a storage organ to a photosynthetically active leaf.
INTRODUCTION
Plastids are major organelles found only in plant and algal cells (Kirk and Tilney-Bassett, 1978). They are responsible for photosynthesis, for the storage of a wide variety of products, and for the synthesis of key molecules required for the maintenance of cellular integrity and activity. Plastids can vary greatly in size, shape, content, and function (Thomson and Whatley, 1980), but little is known about how the remarkable morphological and functional plasticity of plastid development is controlled. The best-studied example of plastid development is the light-induced transformation of etioplasts into chloroplasts in higher plants (Gunning and Steer, 1996). Upon illumination of etiolated seedlings, which lack chlorophyll, major changes occur in the membrane structure and the protein composition of etioplasts (Henningsen and Boynton, 1974). The vast majority of plastid proteins are of nuclear origin, and light-induced control of the transcription of genes that encode these proteins has been regarded as the major mechanism that controls the transition from etioplast to chloroplast (Mullet, 1988). Here, we describe a different strategy used by plants to regulate chloroplast development: the substrate-dependent import of a key enzyme of chlorophyll biosynthesis, the NADPH-protochlorophyllide (Pchlide) oxidoreductase (POR)A.
In Arabidopsis, light-dependent Pchlide reduction is mediated by three structurally related but differentially regulated PORs, denoted PORA, PORB, and PORC (Armstrong et al., 1995; Oosawa et al., 2000; Su et al., 2001). The PORA and PORB genes, but not PORC, are strongly expressed early in seedling development. By contrast, the expression of PORB and PORC, but not PORA, is observed in older seedlings and adult plants. PORA and PORB are both present in etioplasts. Unlike PORB mRNA, the accumulation of PORA mRNA is negatively regulated by light (Apel, 1981; Armstrong et al., 1995). This difference in the expression of PORA and PORB has led to the suggestion that PORA's function in etiolated seedlings might be confined to the initial phase of light-induced chloroplast development (Reinbothe et al., 1995a; Sperling et al., 1997).
A Pchlide-dependent mode of import of PORA had been described for isolated plastids (Reinbothe et al., 1995b), but these results could not be repeated (Aronsson et al., 2000; Dahlin et al., 2000), and they have been attributed to experimental artifacts (Aronsson et al., 2003). In the present work, we have studied the import reactions of PORA and PORB in intact seedlings of Arabidopsis, thus avoiding any alteration of the plastid's integrity and activity that might occur during the isolation and subsequent handling of isolated plastids. We demonstrate that in intact seedlings of Arabidopsis, the import of PORA into plastids of cotyledons is substrate dependent, whereas in true leaves, PORA uptake does not require the presence of Pchlide. This organ specificity of substrate-dependent PORA import reveals a means of controlling plastid protein translocation that in Arabidopsis seems to occur only in cotyledons of seedlings emerging from the dark.
RESULTS
Intracellular Distribution of PORA–Green Fluorescent Protein Fusion Proteins in Light-Adapted and Etiolated Seedlings of Arabidopsis
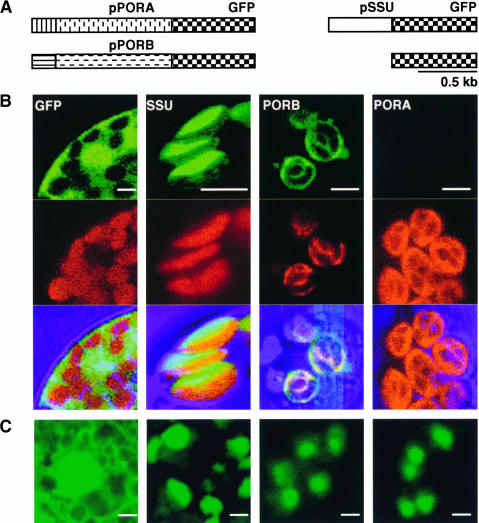
The mode of import of PORA was studied in intact transgenic Arabidopsis plants expressing the precursor of PORA fused to green fluorescent protein (GFP). As controls, plants expressing fusions of precursors of PORB or the small subunit of ribulose-1,5-bisphosphate carboxylase (SSU) with GFP, or GFP alone without a plastid signal sequence attached to it, were analyzed (Figure 1A). The intracellular distribution of the various reporter proteins was first determined in protoplasts isolated from light-adapted green transgenic seedlings. GFP and chlorophyll, as a marker for the thylakoid membrane system of chloroplasts, were detected with the confocal microscope by their bright green and red fluorescence, respectively. PORA-GFP was not detectable in plastids of light-grown seedlings, whereas the other reporter proteins were present (Figure 1B). Merging of the GFP and chlorophyll fluorescence images confirmed the expected localization of PORB and SSU. PORB-GFP was integrated into thylakoid membranes, and SSU-GFP accumulated within the stroma of plastids. Native GFP was excluded from the plastids and accumulated in the surrounding cytoplasm (Figure 1B). Similar results were obtained with intact cotyledons (data not shown). However, in contrast to light-grown seedlings in etiolated plants, not only PORB-GFP and SSU-GFP but also PORA-GFP accumulated within the plastids (Figure 1C). The apparent absence of PORA-GFP from plastids of light-grown seedlings of transgenic plants did not seem to be attributable to an effect on the accumulation of the mRNAs. Specific primer sequences were used to determine the amounts of transcripts of the PORA-GFP and PORB-GFP transgenes in light-grown seedlings by quantitative PCR. As shown in Figure 2, high transcript levels were found in these seedlings, regardless of whether they expressed PORA-GFP or PORB-GFP transgenes, whereas in wild-type control plants, these transcripts were not detectable.
Figure 1.
Intracellular Accumulation of GFP Fusion Derivatives of PORA, PORB, and SSU in Vivo.
(A) Three different cDNA fragments, encoding pPORA-GFP, pPORB-GFP, and pSSU-GFP, were used for the stable transformation of Arabidopsis plants.
(B) and (C) The green fluorescence of GFP and the red fluorescence of chlorophyll were monitored separately using a confocal laser scanning microscope, and then the two fluorescence images were merged. The fluorescence was induced in protoplasts isolated from light-grown seedlings (B) and in cotyledons of etiolated seedlings (C) of transgenic Arabidopsis lines. As a control, the distribution of GFP alone, without a plastid signal sequence attached to it, was monitored. Bars = 5 μm.
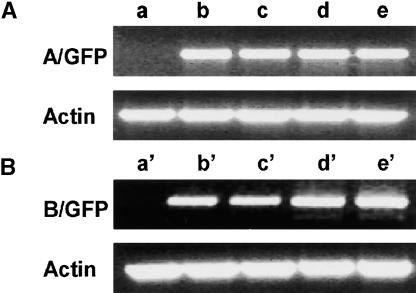
Figure 2.
Expression of PORA-GFP and PORB-GFP Transgenes as Determined by Reverse Transcriptase–Mediated PCR.
(A) PORA-GFP.
(B) PORB-GFP.
Total RNA was extracted from nontransgenic control plants (lanes a and a′) and wild-type plants (lanes b, b′, c, and c′) and flu mutant plants (lanes d, d′, e, and e′) containing transgenes encoding PORA-GFP (lanes b to e) and PORB-GFP (lanes b′ to e′). Plants were either kept under continuous light (lanes a, b, b′, d, and d′) or transferred from the light back to the dark for 6 h (lanes a′, c, c′, e, and e′). Transcript levels of an actin gene were used as an internal amplification control.
Application of Two Noninvasive Methods to Modulate the Endogenous Level of Pchlide in Intact Seedlings
Pigments such as Pchlide and chlorophyll have been reported previously to influence the translocation of plastid proteins (Reinbothe et al., 1995b; Eggink and Hoober, 2000). Etioplasts accumulate higher levels of Pchlide, whereas in chloroplasts, the Pchlide concentration decreases beyond the level of detection; therefore, we suspected that these differences in Pchlide concentration could have contributed to the presence or absence of PORA-GFP in plastids of intact etiolated and light-grown Arabidopsis seedlings, respectively. The biosynthesis of chlorophyll in higher plants relies exclusively on the strictly light-dependent reduction of the immediate precursor of chlorophyll, Pchlide, to chlorophyllide (Beale, 1999). In the dark, stable Pchlide-NADPH-POR ternary complexes are formed, such that absorption of light by the pigment itself leads to its immediate reduction (Griffiths, 1978) (Figure 3). The pigment fraction bound to the POR active site in these ternary complexes thus is termed “photoactive.” The accumulation of additional “nonphotoactive” Pchlide, potentially a lethal sensitizer for photoactive damage upon illumination, normally is prevented by negative feedback control of Glu-tRNA reductase, the first enzyme committed exclusively to tetrapyrrole biosynthesis (Figure 3) (Vothknecht et al., 1998; Meskauskiene and Apel, 2002). Various mutants have been described in which different enzymatic and regulatory steps within this pathway are blocked (von Wettstein et al., 1974; Runge et al., 1995). Two of these mutants of Arabidopsis, xantha2 (Runge et al., 1995) and flu (Meskauskiene et al., 2001), were used in the present study to modulate the endogenous level of Pchlide noninvasively and thus test its possible involvement in regulating the import of PORA into plastids of intact seedlings. The xantha2 mutant is unable to form Mg2+ protoporphyrin IX (Mg2+ProtoIX) from protoporphyrin IX (ProtoIX) and thus lacks Pchlide (Figure 3). Upon supplementation with 5-aminolevulinic acid (ALA), the common precursor of all tetrapyrroles, large amounts of ProtoIX, but no Pchlide, accumulated in etiolated seedlings of homozygous xantha2, whereas in wild-type seedlings treated in the same way, high levels of Pchlide and Mg2+ProtoIX were detected in addition to ProtoIX (Figure 4). Thus, in contrast to other xantha mutants that have been shown to be leaky (Runge et al., 1995), xantha2 seems to be virtually devoid of Pchlide.
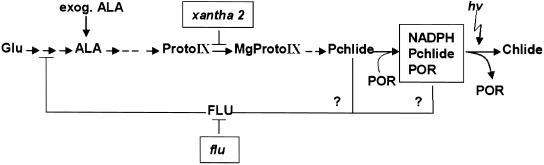
Figure 3.
Simplified Scheme of the FLU-Dependent Control of Chlorophyll Biosynthesis in Higher Plants.
FLU has been identified as a membrane-bound plastid protein that mediates the feedback inhibition of Mg2+ porphyrin biosynthesis by interacting with Glu-tRNA reductase (Meskauskiene et al., 2001; Meskauskiene and Apel, 2002), the first enzyme committed exclusively to tetrapyrrole biosynthesis. As indicated by the question marks, it is not yet known whether Pchlide acts in its free form or as part of the ternary POR-Pchlide-NADPH complex as an effector of feedback inhibition. Inactivation of FLU leads to the overaccumulation of Pchlide in dark-grown flu plants. In the light, continuous photoreduction of Pchlide prevents the accumulation of this pigment in the mutant. Also, feeding of exogenous ALA to dark-grown plants results in the overaccumulation of Pchlide. In the xantha2 mutant of Arabidopsis, the step leading from ProtoIX to Mg2+ProtoIX is blocked, and upon feeding with exogenous ALA, this mutant no longer is able to synthesize detectable amounts of Pchlide in the dark but instead accumulates ProtoIX (Runge et al., 1995) (see text). hv, light.
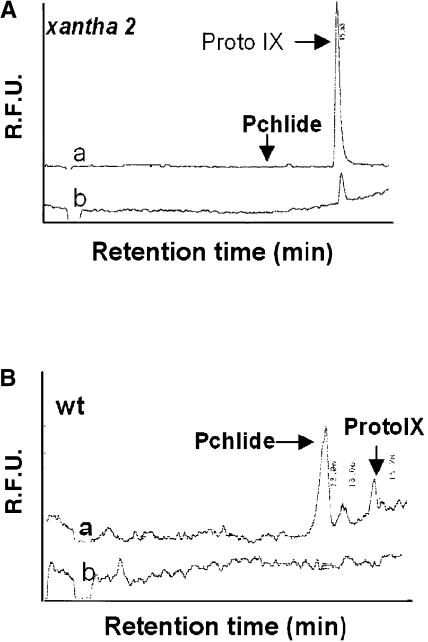
Figure 4.
Biosynthesis of Mg2+ Porphyrins in Wild-Type and Homozygous xantha2 Mutant Seedlings Supplemented with Exogenous ALA.
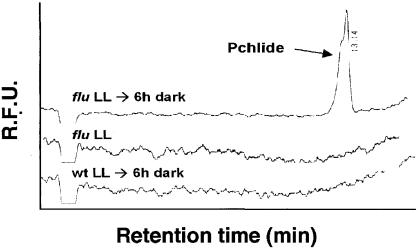
Four-day-old seedlings grown under continuous light were vacuum-infiltrated with 10 mM ALA and subsequently transferred to the dark for 12 h. Porphyrins were extracted and analyzed by HPLC. In samples of xantha2 (A), they were detected by their fluorescence using excitation wavelengths of 404 nm (trace a) and 430 nm (trace b), whereas in wild-type (wt) samples (B), an excitation wavelength of 430 nm was used. In wild-type seedlings (B), large amounts of Pchlide accumulated during the dark incubation with (trace a) but not without (trace b) ALA. In homozygous xantha2 seedlings supplemented with ALA, ProtoIX but not Pchlide was found. R.F.U., relative fluorescence units.
The second mutant, flu, is defective in the metabolic feedback control of ALA synthesis (Figure 3). As a result of this mutation, dark-grown plants no longer are able to restrict the synthesis of Pchlide and thus accumulate a 10- to 15-fold higher amount of this pigment than do wild-type plants (Meskauskiene et al., 2001). When transferred from the dark to the light, homozygous flu mutant seedlings rapidly bleached and died. They could be rescued, however, by germinating the seedlings under constant light. Under these light conditions, Pchlide was photoreduced continuously and its level remained below the level of detection, similar to what has been found in light-grown wild-type plants. After transferring these light-adapted seedlings to the dark, however, flu seedlings rapidly started to accumulate Pchlide, whereas wild-type seedlings failed to do so (Figure 5). Other intermediates of chlorophyll biosynthesis, such as ProtoIX, Mg2+ProtoIX, and Mg2+ProtoIX monomethylester, were barely detectable in either wild-type or mutant plants (data not shown).
Figure 5.
Accumulation of Pchlide in Wild-Type and flu Seedlings.
Wild-type (wt) and flu seedlings were grown for 5 days under continuous light and then transferred to the dark for 6 h. Pchlide was extracted from plants before and after dark treatment and was analyzed by HPLC as described for Figure 4. The fluorescence was measured at 630 nm using an excitation wavelength of 430 nm. In flu seedlings kept under light (LL), no Pchlide was detected, whereas larger amounts of this pigment accumulated during redarkening (LL → 6 h dark). In wild-type seedlings, Pchlide was not detected before or after redarkening. R.F.U., relative fluorescence units.
Effect of Pchlide on the Intracellular Distribution of PORA-GFP in Intact Seedlings of xantha2 and flu
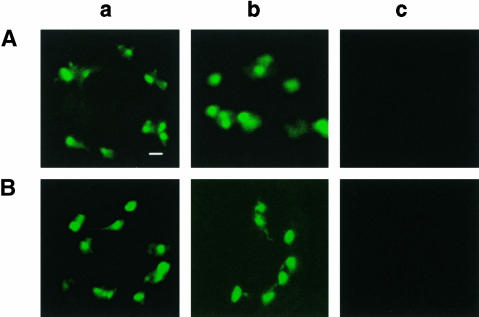
First, the various GFP fusion proteins were expressed stably in the xantha2 mutant of Arabidopsis. Homozygous xantha2 mutants cannot survive beyond the seedling stage. Therefore, heterozygous xantha2 mutants that show the wild-type phenotype were transformed with the various GFP fusion gene constructs. The intracellular distribution of GFP fluorescence was analyzed in 16 etiolated seedlings chosen randomly from the segregating F1 progeny of a transformed heterozygous xantha2 mutant plant and that contained the PORA-GFP construct. In 3 of the 16 seedlings selected, no PORA-GFP signal was detected (Figure 6Ac), whereas the remaining 13 seedlings accumulated large amounts of the protein (data not shown). By contrast, PORB-GFP and SSU-GFP were present in all of the F1 seedlings of heterozygous xantha2 plants (Figures 6Aa and 6Ab). A similar distribution of PORA-GFP also was found in light-grown seedlings (Figure 6B). The genotypes of the 16 plants grown in the dark could be determined easily by transferring them to continuous light. Seedlings of the wild type and heterozygous mutants rapidly turned green, whereas cotyledons of homozygous xantha2 plants retained the yellow color characteristic of etiolated seedlings (Runge et al., 1995). Only the three plants derived from the seedlings that did not accumulate the PORA-GFP fusion protein in the dark turned out to be homozygous xantha2 mutants. All 16 seedlings were derived from the same parent plant and thus had the same genetic background, except for the xantha2 gene. Hence, the failure of homozygous xantha2 mutants to accumulate PORA-GFP must be the result of a block in the chlorophyll biosynthesis pathway and thus may be caused by the lack of Pchlide.
Figure 6.
Intracellular Accumulation of SSU-GFP, PORB-GFP, and PORA-GFP in Etiolated and Light-Grown xantha2 Plants That Lack Pchlide.
(A) Etiolated xantha2.
(B) Light-grown xantha2.
The intracellular distribution of these proteins was monitored by confocal microscopy. SSU-GFP (a) and PORB-GFP (b) accumulated within the plastids under both growth conditions, whereas PORA-GFP (c) was not detectable. Bar = 5 μm.
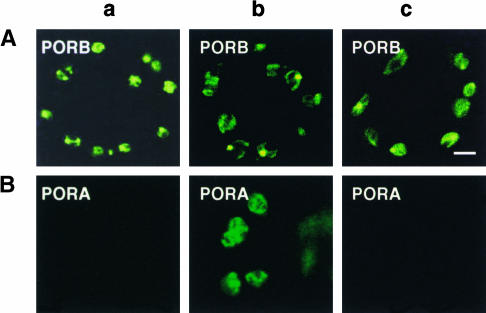
The idea that Pchlide is required for the uptake of PORA into plastids was tested further in a second experiment. By using transgenic flu mutants that express the various GFP fusion proteins, the effect of enhanced levels of Pchlide on the accumulation of PORA in chloroplasts of intact green seedlings also could be analyzed in a noninvasive manner (Figures 5 and 7). In cotyledons of flu and wild-type seedlings kept under continuous light, SSU-GFP (data not shown) and PORB-GFP (Figure 7Aa), but not PORA-GFP (Figure 7Ba), accumulated within chloroplasts. Soon after these seedlings were transferred to the dark, flu started to accumulate Pchlide (Figure 5) and PORA-GFP began to appear in plastids of this mutant (Figure 7Bb), whereas in wild-type control seedlings, the fusion protein was not detected (Figure 7Bc). These differences in PORA-GFP accumulation were not caused by different amounts of transcripts of the transgene in flu and wild-type plants. As shown in Figure 2, similar levels of transcripts of the PORA-GFP and PORB-GFP transgenes were present in these plants.
Figure 7.
Intracellular Accumulation of PORB-GFP and PORA-GFP in flu and in Wild-Type Controls.
(A) PORB-GFP.
(B) PORA-GFP.
Seedlings were grown for 5 days under continuous light (a) and then transferred to the dark for 6 h as described for Figure 5 ([b] and [c]). PORA-GFP accumulates only in chloroplasts of flu plants during the dark period in the presence of Pchlide (Bb), whereas PORB-GFP is present in chloroplasts of both flu and wild-type plants irrespective of the presence or apparent absence of Pchlide ([Aa] to [Ac]). Bar = 5 μm.
The Substrate-Dependent Appearance of PORA in Plastids of Cotyledons Is Not the Result of Changes in Protein Stability
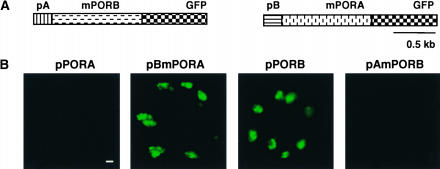
The presence or absence of PORA-GFP in plastids of intact plants is closely correlated with the presence or absence of Pchlide, suggesting that import of the PORA precursor protein requires the presence of the enzyme's substrate. However, as shown previously (Reinbothe et al., 1995a), Pchlide also may have a strong impact on the stability of PORA. POR forms binary and ternary complexes with Pchlide and NADPH that exhibit an enhanced resistance to proteolysis, whereas the apoenzyme is highly susceptible to proteolytic degradation. Hence, the results reported to date do not exclude the possibility that Pchlide may stabilize PORA-GFP in the plastids rather than allow the import of PORA to occur. To distinguish between these two possibilities, chimeric fusion proteins were expressed in transgenic Arabidopsis seedlings. The transit sequences of pPORA and pPORB were exchanged and fused reciprocally to mature PORB-GFP and PORA-GFP, respectively (Figure 8A). When the transit peptide of pPORA (pA) was attached to PORB-GFP, uptake of the fusion protein became dependent on Pchlide, so that it no longer accumulated in the chloroplasts of light-grown seedlings (Figure 8B). Conversely, when linked to the transit peptide of pPORB (pB), PORA-GFP accumulated inside the chloroplasts of green cotyledons (Figure 8B). Thus, the effect of Pchlide on the appearance of PORA-GFP in plastids of intact seedlings is not attributable to changes in protein stability; rather, it reflects the dependence of the import of this protein on the availability of Pchlide.
Figure 8.
Substrate-Dependent Import of pPORA-GFP into Chloroplasts of Cotyledons of Intact Plants.
(A) PORA-GFP and PORB-GFP were expressed in light-grown wild-type seedlings as chimeric fusion proteins in which the transit sequences of pPORA (pA) and pPORB (pB) were exchanged and fused reciprocally to the mature mPORB-GFP and mPORA-GFP, respectively.
(B) The transit peptide of pPORA makes the import of PORB-GFP dependent on Pchlide, such that this fusion protein no longer accumulates in chloroplasts (pAmPORB). Conversely, the signal sequence of pPORB permits the accumulation of PORA-GFP in the chloroplasts in the absence of Pchlide (pBmPORA). As controls, the uptake of nonmodified pPORB-GFP and pPORA-GFP in chloroplasts of light-grown seedlings was monitored (pPORA and pPORB). Bar = 5 μm.
In light-grown seedlings, PORA-GFP not only failed to accumulate within the chloroplasts but also was not detectable outside of the plastids in the surrounding cytoplasm (Figure 1B). This apparent absence of PORA-GFP in light-grown plants devoid of Pchlide may indicate a rapid degradation of the nonimported precursor of PORA, similar to the rapid turnover of other precursor proteins that had been reported previously (Apel and Kloppstech, 1980; Bennett, 1981; Bellemare et al., 1982; Reid and Schatz, 1982), or may be caused by the Pchlide-dependent control of PORA-GFP mRNA translation.
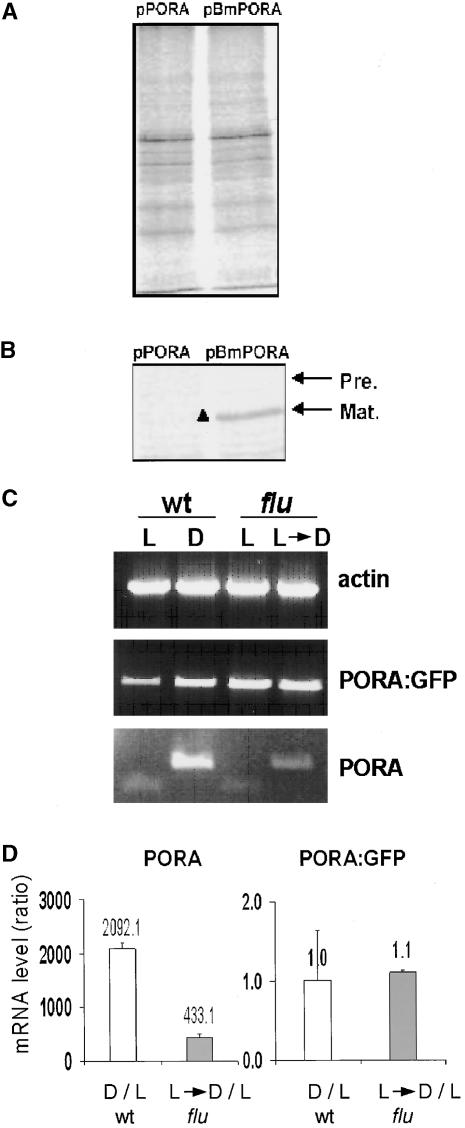
These proposed modes of control of PORA biosynthesis and accumulation were tested in two different ways. First, proteins were pulse labeled with 35S-Met in light-adapted transgenic Arabidopsis seedlings that expressed either pPORA-GFP or chimeric PORA-GFP, in which the transit peptide of PORA had been replaced by the transit peptide of PORB (pBmPORA) (Figure 8). Total protein extracts from both plant samples containing equal amounts of incorporated radioactivity were either separated directly electrophoretically (Figure 9A) or used for the immunoprecipitation of PORA-GFP (Figure 9B). In plants transformed with the pPORA-GFP construct, no radioactively labeled PORA-GFP was detected, whereas in plants containing the chimeric pBmPORA construct, accumulation of the mature protein was demonstrated by immunoprecipitation (Figure 9B).
Figure 9.
Effect of Pchlide on the Biosynthesis and Accumulation of PORA-GFP in Transgenic Arabidopsis Seedlings.
(A) and (B) Pulse labeling of PORA-GFP in transgenic plants expressing pPORA-GFP (pPORA) or a chimeric PORA-GFP protein in which the signal peptide of PORA had been replaced by the signal sequence of PORB (pBmPORA). Total protein extracts from cotyledons of both plants containing equal amounts of radioactivity were either separated electrophoretically (A) or used for the immunoprecipitation of PORA-GFP, and the immunoprecipitation was subsequently dissolved and separated electrophoretically (B). Radioactively labeled proteins were detected by autoradiography. Arrows mark the positions of the mature (Mat.) PORA-GFP (black triangle) and its nonprocessed precursor protein (Pre.).
(C) Relative concentrations of PORA-GFP mRNAs associated with polysomes of light-grown (L) and etiolated (D) wild-type seedlings (wt) and light-grown flu seedlings (flu) either kept under continuous light (L) or transferred to the dark for 6 h before harvest (L→D). As a control, the concentrations of endogenous PORA mRNA were determined within these fractions. Transcript levels of an actin gene were used as an internal amplification control.
(D) The relative concentrations of mRNAs as revealed in (C) were confirmed by real-time PCR. Numbers above each column indicate the ratios of either PORA or PORA-GFP mRNAs of etiolated versus light-grown wild-type seedlings (white columns) and light-grown flu seedlings that had been returned to the dark for 6 h versus light-grown flu seedlings (gray columns). Results shown are means ± sd of four measurements.
Second, the association of PORA-GFP mRNAs with polysomes was analyzed in etiolated and light-grown 4-day-old wild-type seedlings and in flu seedlings of similar age that were kept under continuous light or transferred to the dark for 6 h. As a control, the levels of endogenous PORA mRNA also were determined (Figures 9C and 9D). Upon illumination of etiolated Arabidopsis seedlings, the PORA mRNA concentration declines rapidly, whereas in light-grown seedlings that are transferred back to the dark, PORA mRNA starts to reaccumulate (Armstrong et al., 1995; Su et al., 2001). The relative concentrations of PORA-GFP and PORA mRNAs were determined by PCR. The levels of PORA-GFP mRNAs associated with polysomes were similar in all four plant samples, regardless of whether these seedlings accumulated higher amounts of Pchlide or were devoid of this pigment (Figure 9C), whereas PORA mRNAs were found only within the polysomal fractions of etiolated seedlings and, to a lesser extent, of light-grown flu seedlings that had been transferred back to the dark. In light-grown wild-type and flu seedlings, these mRNAs were not detectable (Figure 9C). These results could be confirmed by real-time PCR. Equal amounts of PORA-GFP mRNAs were present within the polysomal fractions of etiolated and light-grown wild-type seedlings and light-grown flu seedlings with or without a subsequent dark treatment. However, the corresponding levels of PORA mRNA in etiolated wild-type and light-grown flu seedlings returned to the dark exceeded those in light-grown wild-type and flu seedlings by factors of ∼2000 and 430, respectively (Figure 9D). The results of these two experiments do not support the Pchlide-dependent control of PORA-GFP mRNA translation; rather, they suggest that in the absence of Pchlide, nonimported preproteins of PORA-GFP are degraded rapidly.
Proteins that are subject to rapid degradation have been shown previously to confer this instability also upon GFP moieties fused to these proteins (Clute and Pines, 1999), whereas native GFP alone seems to be well protected against proteolytic attack (Zhong et al., 2003) (Figure 1).
The Pchlide-Dependent Import of PORA-GFP in Plastids of Intact Seedlings Is Confined to the Cotyledons and Does Not Occur in True Leaves
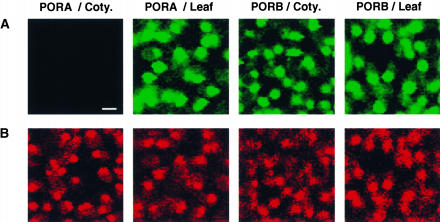
To date, the substrate-dependent import of PORA-GFP has been shown to operate in cotyledons. PORA expression is not only negatively controlled by light but also is subject to developmental control. In contrast to young seedlings at the cotyledon stage, older seedlings and chlorophyll-containing organs of adult plants do not seem to express this protein (Armstrong et al., 1995). Thus, it was of interest to determine whether the substrate dependence of PORA import was maintained in tissues that normally would not be expected to synthesize and translocate the PORA precursor into chloroplasts. The distributions of PORA-GFP and PORB-GFP in cotyledons and primary leaves of light-grown transgenic Arabidopsis seedlings were compared. In control plants expressing the PORB-GFP construct, both types of leaves accumulated similarly high levels of this protein (Figure 10). PORA-GFP did not accumulate in green cotyledons, whereas in primary leaves of the same plants, large amounts of this protein were present in chloroplasts (Figure 10), although Pchlide levels in such leaves were below the level of detection. Thus, the substrate dependence of PORA import is subject to developmental control and is restricted to cotyledons in which this protein is expressed normally.
Figure 10.
Organ-Specific Import of PORA-GFP into Chloroplasts of 5-Day-Old Transgenic Wild-Type Seedlings of Arabidopsis.
PORA-GFP accumulates in chloroplasts of the first true leaf (PORA/Leaf) but not in chloroplasts of cotyledons (PORA/Coty.) of light-grown seedlings. PORB-GFP accumulates in chloroplasts of both types of leaves (PORB/Leaf and PORB/Coty.). The green fluorescence of GFP (A) and the red fluorescence of chlorophyll (B) were monitored by confocal microscopy. Bar = 5 μm.
DISCUSSION
Cotyledons of etiolated Arabidopsis seedlings emerging from the dark undergo a dramatic light-induced transformation from being storage organs in the dark to becoming the first photosynthetically active leaves, which defines the transition from heterotrophic to autotrophic growth of these plants. Most of the changes that occur during the light-dependent derepression of the photomorphogenesis program are under the control of phytochrome (Quail et al., 1995), with the exception of chlorophyll formation, which in one of its final steps, catalyzed by POR, requires direct activation by light. Among the three POR proteins of Arabidopsis, PORA is the only one whose activity seems to be confined to the initial phase of light-induced chloroplast formation. In addition to its involvement in the photoreduction of Pchlide to chlorophyllide, PORA also has been implicated in protecting seedlings against photooxidative damage (Sperling et al., 1997). Although PORA is abundantly present in etioplasts, it disappears selectively soon after the onset of illumination. It is interesting that this negative regulation of PORA by light is mediated simultaneously at various levels. First, the concentration of PORA mRNA declines drastically during the illumination of dark-grown seedlings (Apel, 1981). Second, PORA becomes destabilized within the plastids soon after the start of illumination and is degraded by a light-induced protease (Reinbothe et al., 1995a). Finally, as shown here, the plastid's ability to import the PORA precursor is reduced drastically during the transition from etioplast to chloroplast. This effect is the result of the rapid decline in the Pchlide content of plastids in the cotyledons of illuminated seedlings.
The substrate-dependent control of PORA import may serve a dual purpose. At least part of the Pchlide is synthesized within the plastid envelope (Pineau et al., 1993). The suggested interaction between Pchlide and the PORA precursor during the translocation step might help to ensure the immediate assembly of the ternary photoactive PORA-NADPH-Pchlide complex, which, in contrast to the PORA apoenzyme, is very stable and well protected against proteolytic attack (Reinbothe et al., 1995a). At the same time, it may prevent the accumulation of potentially deleterious free tetrapyrrole intermediates, such as Pchlide, that can act as photosensitizers in the light and cause photooxidative damage in the plastid compartment (Meskauskiene et al., 2001).
This protective mechanism would operate efficiently only if an excess of nonprocessed PORA precursor protein were available outside of the plastid compartment. Indeed, trace amounts of PORA precursor proteins have been shown to be bound to the outer surface of the envelope membranes of isolated etiochloroplasts in a transport-competent conformation (Joyard et al., 1990; Reinbothe et al., 1996). As shown here by the labeling and immunoprecipitation of PORA-GFP, in intact plants, nonimported preproteins of PORA did not accumulate to detectable levels. One could envisage two different mechanisms by which plants could avoid such an overaccumulation of pPORA: rapid turnover of the nonimported precursor proteins in the cytoplasm, and/or translational control of PORA gene expression that is coupled to the import block of pPORA caused by the lack of Pchlide.
Rapid turnover of nonimported precursors of PORA would be expected to become effective only during the illumination of predarkened plants, when the concentration of Pchlide declines rapidly and this pigment soon becomes a limiting factor for the import reaction. At the same time, the concentration of PORA mRNA declines drastically (Apel, 1981), thus avoiding a futile cycle of PORA synthesis in illuminated plants. Whether this light-dependent regulation of PORA biosynthesis also includes an additional Pchlide-dependent control of PORA mRNA translation is not known, but this seems less likely. Pchlide is confined to the plastid compartment (op den Camp et al., 2003), and it is generally accepted that nucleus-encoded plastid proteins are synthesized on free cytosolic polysomes that are not attached to the outer surfaces of plastids. Thus, the physical contact between Pchlide and the nascent pPORA signal sequence that would be required for such a Pchlide-dependent control of pPORA mRNA translation seems highly unlikely. Furthermore, the association of PORA-GFP mRNAs with polysomes in transgenic Arabidopsis plants was not altered visibly during the illumination of etiolated seedlings after the Pchlide concentration had decreased beyond the level of detection or in flu seedlings after Pchlide accumulation had been induced by transferring these plants from the light to the dark.
The observed differences in the import of PORA between cotyledons and primary leaves must be the result of differences in the import mechanisms that operate in these two types of leaves. Based mainly on studies of isolated chloroplasts, a general import pathway has been identified that involves multimeric complexes in the outer and inner chloroplast envelope membranes, designated Toc and Tic complexes, respectively (Schnell et al., 1997). Although this general import pathway initially was considered to be engaged in the translocation of all plastid proteins (Jarvis and Soll, 2001), the discovery of multiple variants of subunits of the Toc and Tic homologs in Arabidopsis has led to the suggestion that several import routes into chloroplasts exist that might be involved in the import of distinct subsets of plastid proteins (Jarvis et al., 1998; Bauer et al., 2000). There are conflicting reports regarding whether or not pPORA is imported into isolated chloroplasts by the general Tic-Toc machinery (Aronsson et al., 2000) or by a separate import channel (Reinbothe et al., 2000). A Pchlide-dependent mode of import of PORA had been described previously for isolated plastids (Reinbothe et al., 1995b), but these results could not be repeated and confirmed (Aronsson et al., 2000; Dahlin et al., 2000; Q. Su and K. Apel, unpublished results), and they have since been attributed to experimental artifacts (Aronsson et al., 2003).
Some of the import studies in which Pchlide was shown not to be required for the import of PORA were performed with isolated pea chloroplasts (Dahlin et al., 2000). Pea plants express only one POR gene and apparently lack PORA (Spano et al., 1992). Their cotyledons are used exclusively as storage organs and do not undergo the light-dependent transformation into photosynthetically active leaves as in other dicotyledonous plants, such as Arabidopsis. This difference in seedling development might explain why chloroplasts isolated from pea seedlings do not show a Pchlide-dependent import of PORA. However, similar import studies also were performed with chloroplasts isolated from barley (Dahlin et al., 2000; Aronsson et al., 2003), and again, no Pchlide requirement for the import of PORA was found. On the other hand, Reinbothe et al. (2000) reported Pchlide-dependent PORA uptake for plastids isolated not only from barley but also from other monocotyledonous and dicotyledonous plants, including pea and Arabidopsis. Different protocols that had been used in these two sets of experiments might explain the discrepancies between their findings. The substrate-dependent import of PORA into isolated chloroplasts of Arabidopsis reported by Reinbothe et al. (2000) could not be confirmed by our present in vivo study. The in vitro import studies were performed with chloroplasts prepared from mature leaves (Reinbothe et al., 2000) that in intact plants do not show the Pchlide requirement for PORA uptake (Figure 10).
Import studies with isolated chloroplasts have been extremely successful in identifying and characterizing the various constituents of the membrane complexes that facilitate the direct translocation of preproteins from the cytoplasm (Jarvis and Soll, 2001). However, they seem to be less suitable for assaying the activity of additional factors that confer organ specificity and substrate dependence of import, the features that characterize the import of PORA seen in the present work. It seems likely that in vivo studies using either wild-type or mutant plants that are deficient in specific subunits of the import channels will be needed to discover novel and divergent import characteristics not only for PORA and PORB but possibly also for other sets of plastid proteins.
METHODS
Plant Materials
For the cultivation of mature plants, seeds of Arabidopsis thaliana ecotype Columbia (Col-0) were sown on soil and the plants were grown under a 16-h-light/8-h-dark cycle (80 to 110 μmol·m−2·s−1). flu plants were kept under continuous light. Seedlings were cultivated by germinating surface-sterilized seeds on plates of Murashige and Skoog (1962) (MS) agar, in some cases supplemented with 0.5% sucrose. Plated seeds were kept in the dark for 4 days, or moved to a 16-h-light/8-h-dark cycle for 4 to 12 days, or, in the case of the flu mutant, kept under continuous light (80 to 100 μmol·m−2·s−1) for 5 days and then transferred for up to 6 h to the dark. Protoplasts were isolated according to Danon and Gallois (1998).
Detection of Transgene Expression in Arabidopsis Seedlings by Reverse Transcriptase–Mediated PCR
Total RNA was extracted from 4-day-old transgenic seedlings, and reverse transcriptase–mediated (RT) PCR was performed as described by Melzer et al. (1999). Transcripts of the transgenes were detected using PORA and PORB gene sequences at position +915 bp from the start codon as the forward primer and the GFP sequence at position +460 bp from the start codon as the reverse primer.
Extraction and Measurement of Tetrapyrroles
Tetrapyrroles were extracted with 90% acetone and 10% 0.1 M NH4OH in water. Porphyrins were separated by HPLC on a C18 reverse-phase silica gel column (Nucleosil ODS 5 μm, 250 × 4.6 mm; Machery Nagel, Oensingen, Switzerland) and were detected by their fluorescence using the following excitation and emission (ex/em) wavelengths: 430/630 nm for Pchlide, 416/594 nm for Mg2+ProtoIX and Mg2+ProtoIX monomethylester, and 404/630 nm for ProtoIX (La Rocca et al., 2001).
Construction and Detection of the GFP Fusion Proteins in Vivo
The plasmid DNAs carrying the PORA and PORB cDNAs of Arabidopsis (Armstrong et al., 1995) were modified by inserting internal EcoRI and NcoI recognition sites upstream of the start codon and downstream of the stop codon, respectively. The PCR products were cut with EcoRI and NcoI and ligated into the multicloning site of the pSH9 vector between the 35S promoter and the terminal polyadenylation site. The EGFP cDNA was excised from the pCL60 plasmid (Clontech, Palo Alto, CA) with NcoI and inserted into the modified pSH9 plasmid containing the PORA and PORB cDNAs. Competent Escherichia coli cells (DH5α) were transformed with this plasmid. After propagation, the amplified cDNA was isolated and partially digested with HindIII. The fragments containing the 35S promoter, Ω element, PORA or PORB, EGFP, and the terminal polyadenylation site were ligated into the binary vector pCAMBIA 3300 containing a phosphinothricin gene as a selectable marker in planta.
To clone the small subunit of ribulose-1,5-bisphosphate carboxylase (SSU), total RNA was isolated from 4-day-old Arabidopsis seedlings (Col-0). The isolation of total RNA and the subsequent RT-PCR procedure were based on the protocol of Melzer et al. (1990). The cDNA encoding the precursor of SSU was amplified from the total cDNA. The construction of the DNA encoding SSU-GFP was performed as described for PORA-GFP and PORB-GFP. For the construction of cDNAs encoding chimeric pAmPORB and pBmPORA, the signal sequences of PORA and PORB and the sequences of the mature proteins were amplified separately. The cDNA encoding the signal sequence of PORA was ligated with the cDNA encoding the mature PORB (pAmPORB) or vice versa (pBmPORA). These chimeric cDNAs were ligated with the EGFP cDNA and finally inserted into the binary vector as described above.
Competent cells of Agrobacterium tumefaciens C58 were transformed with the various vectors and then used for stable in planta transformation of the wild type, the flu mutant, or heterozygous xantha2 mutants of Arabidopsis, all ecotype Col-0. For stable transformation, flu plants were kept under continuous light, whereas the other plants were grown under a 16-h-light/8-h-dark cycle. Primary bolts were removed to induce the growth of secondary bolts. For each construct, 10 plants were transformed when secondary bolts reached a length of 10 to 15 cm. A. tumefaciens–mediated transformation of Arabidopsis was performed as described by Bechtold et al. (1993). Transgenic plants were selected on MS agar plates containing phosphinothricin (25 mg/L). The green fluorescence of GFP and the red fluorescence of chlorophyll were monitored using a confocal laser scanning microscope (TCS-NT; Leica Microsystems, Heidelberg, Germany) with Kr/Ar laser excitation. GFP and chlorophyll fluorescence were induced at an excitation wavelength of 488 nm. GFP and chlorophyll were detected at emission wavelengths of 507 to 520 nm and 620 to 700 nm, respectively. TCS-NT software version 1.6.587 (Leica Microsystems) and Adobe Photoshop 5.5 (Adobe Systems, San Jose, CA) were used for image acquisition and processing, respectively.
Pulse Labeling of PORA-GFP in Vivo
Arabidopsis seedlings used for pulse labeling were germinated on MS medium under continuous light for 4 days, and five pairs of cotyledons were incubated with 10 mM sodium phosphate buffer, pH 7.0, containing 50 μCi of 35S-Met for 1 h. Subsequent steps were performed according to Barkan et al. (1994) and McCormac and Barkan (1999). Total protein was solubilized and subjected to immunoprecipitation with polyclonal anti-GFP (Living Colors Full-Length A.v. Polyclonal Antibody; Clontech). A total of 2,000,000 incorporated cpm was used for each immunoprecipitation. Fusion proteins bound to antisera were pulled down by protein A–Sepharose CL-4B (Amersham Biosciences). Immunoprecipitated proteins were fractionated on 10% SDS-polyacrylamide gels and detected by autoradiography.
Analysis of Polysomal PORA-GFP mRNAs
Polysomes were isolated from etiolated and light-grown 4-day-old transgenic seedlings that expressed PORA-GFP and from transgenic flu plants that expressed the same fusion protein; they were kept for 4 days under continuous light with an additional final 6-h dark or light treatment, respectively. Seedlings were frozen in liquid nitrogen and ground to a fine powder with a mortar and pestle. One milliliter of polysome extraction buffer was added to 0.2 g fresh weight as described previously (Barkan, 1998). Nuclei and insoluble cellular debris were removed by two centrifugation steps each for 5 min at 14,000 rpm in an Eppendorf microfuge. The resulting supernatant was centrifuged at 55,000 rpm for 50 min in a Beckman ultracentrifuge (Optima, TLA 120.2 rotor). The pellets were dried at room temperature and dissolved, and the RNAs were purified by phenol extraction and ethanol precipitation (Barkan, 1998). One microgram of each RNA sample was used for cDNA synthesis, and similar dilution steps of cDNAs were used for the PCR analysis of each sample to ensure a correlation between signal and transcript abundance. For the amplification of pPORA-GFP transcripts, a pPORA gene-specific sequence (5′-ATTTCACTTTCGGAGCA-3′) was used as the forward primer and a GFP gene-specific sequence (5′-TCTTCTGCTTGTCGGCCATG-3′) was used as the reverse primer. Endogenous PORA transcripts were detected by PCR using the PORA-specific sequences 5′-CATTACACTCTTTAAGTCTTAAACG-3′ as the forward primer and 5′-AACCACGTTTCCTTTTCTAAGT-3′ as the reverse primer. The following PCR conditions were used: predenaturation at 94°C for 2 min; 30 cycles of 30 s at 94°C, 30 s at 56°C, and 2 min at 72°C; a last extension for 7 min at 72°C; and hold at 10°C. Quantitative real-time PCR analysis was performed with the ABI PRISM 7700 Sequence Detection System (Applied Biosystems, Foster City, CA). RNAs were treated with RQ1 RNase-Free DNase (Promega, Madison, WI) and reverse transcribed using random hexamers and SuperScript II RNase H− Reverse Transcriptase (Invitrogen, Carlsbad, CA) according to each manufacturer's recommendations.
5-Aminolevulinic Acid Feeding Experiments
Four-day-old light-grown homozygous xantha2 and wild-type seedlings were vacuum-infiltrated with 10 mM 5-aminolevulinic acid–KOH, pH 7.9, for 3 min and transferred to the dark for 12 h. Acetone extracts of these seedlings were prepared under green safelight and used for HPLC analysis to detect the relative amounts of ProtoIX and Pchlide.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Klaus Apel, klaus.apel@ipw.biol.ethz.ch.
Accession Number
The GenBank accession number for the small subunit of ribulose-1,5-bisphosphate carboxylase is CAA31948.
Acknowledgments
This article is dedicated to Lawrence Bogorad. We are indebted to Dieter Rubli for art work, to Martha Geier-Bächtold for editorial work, to Paul Hardy for correcting the language, and to Quingxiang Su for his support. This work was supported by the Swiss Federal Institute of Technology and the Swiss National Science Foundation.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.015008.
References
- Apel, K. (1981). The protochlorophyllide holochrome of barley (Hordeum vulgare L.): Phytochrome-induced decrease of translatable mRNA coding for the NADPH-protochlorophyllide oxidoreductase. Eur. J. Biochem. 120, 89–93. [DOI] [PubMed] [Google Scholar]
- Apel, K., and Kloppstech, K. (1980). The effect of light on the biosynthesis of the light-harvesting chlorophyll a/b protein: Evidence for the requirement of chlorophyll a for the stabilization of the apoprotein. Planta 150, 426–430. [DOI] [PubMed] [Google Scholar]
- Armstrong, G.A., Runge, S., Frick, G., Sperling, U., and Apel, K. (1995). Identification of NADPH:protochlorophyllide oxidoreductases A and B: A branched pathway for light-dependent chlorophyll biosynthesis in Arabidopsis thaliana. Plant Physiol. 108, 1505–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aronsson, H., Sohrt, K., and Soll, J. (2000). NADPH:protochlorophyllide oxidoreductase uses the general import route into chloroplasts. Biol. Chem. 381, 1263–1267. [DOI] [PubMed] [Google Scholar]
- Aronsson, H., Sundqvist, C., and Dahlin, C. (2003). POR hits the road: Import and assembly of a plastid protein. Plant Mol. Biol. 51, 1–9. [DOI] [PubMed] [Google Scholar]
- Barkan, A. (1998). Approaches to investigating nuclear genes that function in chloroplast biogenesis in land plants. Methods Enzymol. 297, 38–57. [Google Scholar]
- Barkan, A., Walker, M., Nolasco, M., and Johnson, D. (1994). A nuclear mutation in maize blocks the processing and translation of several chloroplast messenger RNAs and provides evidence for the differential translation of alternative messenger RNA forms. EMBO J. 13, 3170 3181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer, J., Chen, K., Hiltbrunner, A., Wehrli, E., Eugster, M., Schnell, D., and Kessler, F. (2000). The major protein import receptor of plastids is essential for chloroplast biogenesis. Nature 403, 203–207. [DOI] [PubMed] [Google Scholar]
- Beale, S.I. (1999). Enzymes of chlorophyll biosynthesis. Photosynth. Res. 60, 43–73. [Google Scholar]
- Bechtold, N., Ellis, J., and Pelletier, G. (1993). In planta Agrobacterium-mediated gene transfer by infiltration of adult Arabidopsis thaliana plants. C. R. Acad. Sci. Paris 316, 1194–1199. [Google Scholar]
- Bellemare, G., Bartlett, S., and Chua, N.-H. (1982). Biosynthesis of chlorophyll a/b-binding polypeptides in wild type and the chlorina f2 mutant of barley. J. Biol. Chem. 257, 7762–7767. [PubMed] [Google Scholar]
- Bennett, J. (1981). Biosynthesis of the light-harvesting chlorophyll a/b protein: Polypeptide turnover in darkness. Eur. J. Biochem. 118, 61–70. [DOI] [PubMed] [Google Scholar]
- Clute, P., and Pines, J. (1999). Temporal and spatial control of cyclin B1 destruction in metaphase. Nat. Cell Biol. 1, 82–87. [DOI] [PubMed] [Google Scholar]
- Dahlin, C., Aronsson, H., Almkvist, J., and Sundqvist, C. (2000). Protochlorophyllide-independent import of two NADPH:Pchlide oxidoreductase proteins (PORA and PORB) from barley into isolated plastids. Physiol. Plant. 109, 298–303. [Google Scholar]
- Danon, A., and Gallois, P. (1998). UV-C radiation induces apoptotic-like changes in Arabidopsis thaliana. FEBS Lett. 437, 131–136. [DOI] [PubMed] [Google Scholar]
- Eggink, L.L., and Hoober, J.K. (2000). Chlorophyll binding to peptide maquettes containing a retention motif. J. Biol. Chem. 275, 9087–9090. [DOI] [PubMed] [Google Scholar]
- Griffiths, W.T. (1978). Reconstitution of chlorophyllide formation by isolated etioplast membranes. Biochem. J. 174, 681–692. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gunning, B.E.S., and Steer, M.W. (1996). Plant Cell Biology: Structure and Function. (Sudbury, MA: Jones and Bartlett).
- Henningsen, K.W., and Boynton, J.E. (1974). Macromolecular physiology of plastids. IX. Development of plastid membranes during greening of dark-grown barley seedlings. J. Cell Sci. 15, 31–35. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., Chen, L.-J., Li, H.-M., Peto, C.A., Fankhauser, F., and Chory, J. (1998). An Arabidopsis mutant defective in the plastid general protein import apparatus. Science 282, 100–103. [DOI] [PubMed] [Google Scholar]
- Jarvis, P., and Soll, J. (2001). Toc, Tic and chloroplast import. Biochim. Biophys. Acta 1541, 64–79. [DOI] [PubMed] [Google Scholar]
- Joyard, J., Block, M., Pineau, B., Albrieux, C., and Douce, R. (1990). Envelope membranes from mature spinach chloroplasts contain a NADPH:protochlorophyllide reductase on the cytosolic side of the outer membrane. J. Biol. Chem. 265, 21820–21827. [PubMed] [Google Scholar]
- Kirk, J.T., and Tilney-Bassett, R.A.E. (1978). The Plastids. (Amsterdam: Elsevier).
- La Rocca, N., Rascio, N., Oster, U., and Rüdiger, W. (2001). Amitrole treatment of etiolated barley seedlings leads to deregulation of tetrapyrrole synthesis and to reduced expression of Lhc and RbcS genes. Planta 213, 101–108. [DOI] [PubMed] [Google Scholar]
- McCormac, D.J., and Barkan, A. (1999). A nuclear gene in maize required for the translation of the chloroplast atpB/E mRNA. Plant Cell 11, 1709–1716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Melzer, S., Kampmann, G., Chandler, J., and Apel, K. (1999). FPF1 modulates the competence to flowering in Arabidopsis. Plant J. 18, 395–405. [DOI] [PubMed] [Google Scholar]
- Melzer, S., Majewski, D.M., and Apel, K. (1990). Early changes in gene expression during the transition from vegetative to generative growth in the long-day plant Sinapis alba. Plant Cell 2, 953–961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meskauskiene, R., and Apel, K. (2002). Interaction of FLU, a negative regulator of tetrapyrrole biosynthesis, with the glutamyl-tRNA reductase requires the tetratricopeptide repeat domain of FLU. FEBS Lett. 532, 27–30. [DOI] [PubMed] [Google Scholar]
- Meskauskiene, R., Nater, M., Goslings, D., Kessler, F., op den Camp, R., and Apel, K. (2001). FLU: A negative regulator of chlorophyll biosynthesis in Arabidopsis thaliana. Proc. Natl. Acad. Sci. USA 98, 12826–12831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mullet, J.E. (1988). Chloroplast development and gene expression. Annu. Rev. Plant Physiol. Plant Mol. Biol. 39, 475–502. [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Oosawa, N., Masuda, T., Awai, K., Fusada, N., Shimada, H., Ohta, H., and Takamiya, K. (2000). Identification and light-induced expression of a novel gene of NADPH-protochlorophyllide oxidoreductase isoform in Arabidopsis thaliana. FEBS Lett. 474, 133–136. [DOI] [PubMed] [Google Scholar]
- op den Camp, R., Przybyla, D., Ochsenbein, C., Laloi, C., Kim, C., Danon, A., Wagner, D., Hideg, E., Göbel, C., Feussner, I., Nater, M., and Apel, K. (2003). Rapid induction of distinct stress responses after the release of singlet oxygen in Arabidopsis. Plant Cell 15, 2320–2332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pineau, B., Gerard-Hirne, C., Douce, R., and Joyard, J. (1993). Identification of the main species of tetrapyrrole pigments in envelope membranes from spinach chloroplasts. Plant Physiol. 102, 821–828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quail, P.H., Boylan, M.T., Parks, B.M., Short, T.W., Xu, Y., and Wagner, D. (1995). Phytochrome: Photosensory perception and signal transduction. Science 268, 675–680. [DOI] [PubMed] [Google Scholar]
- Reid, G.A., and Schatz, G. (1982). Import of proteins into mitochondria: Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J. Biol. Chem. 257, 13056–13061. [PubMed] [Google Scholar]
- Reinbothe, S., Mache, R., and Reinbothe, C. (2000). A second, substrate-dependent site of protein import into chloroplasts. Proc. Natl. Acad. Sci. USA 97, 9795–9800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe, S., Reinbothe, C., Holtorf, H., and Apel, K. (1995. a). Two NADPH:protochlorophyllide oxidoreductases in barley: Evidence for the selective disappearance of PORA during the light-induced greening of etiolated seedlings. Plant Cell 7, 1933–1940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe, S., Reinbothe, C., Neumann, D., and Apel, K. (1996). A plastid enzyme arrested in the step of precursor translocation in vivo. Proc. Natl. Acad. Sci. USA 93, 12026–12030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reinbothe, S., Runge, S., Reinbothe, C., van Cleve, B., and Apel, K. (1995. b). Substrate-dependent transport of the NADPH:protochlorophyllide oxidoreductase into isolated plastids. Plant Cell 7, 161–172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Runge, S., van Cleve, B., Lebedev, N., Armstrong, G.A., and Apel, K. (1995). Isolation and classification of chlorophyll-deficient xantha mutants of Arabidopsis thaliana. Planta 197, 490–500. [DOI] [PubMed] [Google Scholar]
- Schnell, D., Blobel, G., Keegstra, K., Kessler, F., Ko, K., and Soll, J. (1997). A consensus nomenclature for the protein import components of the chloroplast envelope. Trends Cell Biol. 7, 303–304. [DOI] [PubMed] [Google Scholar]
- Spano, A.J., He, Z., Michel, H., Hunt, D.F., and Timko, M.P. (1992). Molecular cloning, nuclear gene structure and developmental expression of NADPH:protochlorophyllide oxidoreductase in pea (Pisum sativum L.). Plant Mol. Biol. 18, 967–972. [DOI] [PubMed] [Google Scholar]
- Sperling, U., van Cleve, B., Frick, G., Apel, K., and Armstrong, G.A. (1997). Overexpression of light-dependent PORA or PORB in plants depleted of endogenous POR by far-red light enhances seedling survival in white light and protects against photooxidative damage. Plant J. 12, 649–658. [DOI] [PubMed] [Google Scholar]
- Su, Q., Frick, G., Armstrong, G.A., and Apel, K. (2001). POR C of Arabidopsis thaliana: A third light- and NADPH-dependent protochlorophyllide oxidoreductase that is differentially regulated by light. Plant Mol. Biol. 47, 805–813. [DOI] [PubMed] [Google Scholar]
- Thomson, W.W., and Whatley, J.M. (1980). Development of nongreen plastids. Annu. Rev. Plant Physiol. 31, 375–394. [Google Scholar]
- von Wettstein, D., Kahn, A., Nielsen, O.F., and Gough, S.P. (1974). Genetic regulation of chlorophyll synthesis analyzed with mutants of barley. Science 184, 800–802. [DOI] [PubMed] [Google Scholar]
- Vothknecht, U.C., Kannangara, C.G., and von Wettstein, D. (1998). Barley glutamyl tRNA Glu reductase: Mutations affecting haem inhibition and enzyme activity. Phytochemistry 47, 513–519. [DOI] [PubMed] [Google Scholar]
- Zhong, R., Wan, J., Jin, R., and Lamppa, G. (2003). A pea antisense gene for the chloroplast stromal processing peptidase yields seedling lethals in Arabidopsis: Survivors show defective GFP import in vivo. Plant J. 34, 802–812. [DOI] [PubMed] [Google Scholar]