Abstract
Jasmonic acid (JA) is a fatty acid–derived signaling molecule that regulates a broad range of plant defense responses against herbivores and some microbial pathogens. Molecular genetic studies in Arabidopsis have established that JA also performs a critical role in anther and pollen development but is not essential for other developmental aspects of the plant's life cycle. Here, we describe the phenotypic and molecular characterization of a sterile mutant of tomato (jasmonic acid–insensitive1 [jai1]) that is defective in JA signaling. Although the mutant exhibited reduced pollen viability, sterility was caused by a defect in the maternal control of seed maturation, which was associated with the loss of accumulation of JA-regulated proteinase inhibitor proteins in reproductive tissues. jai1 plants exhibited several defense-related phenotypes, including the inability to express JA-responsive genes, severely compromised resistance to two-spotted spider mites, and abnormal development of glandular trichomes. We demonstrate that these defects are caused by the loss of function of the tomato homolog of CORONATINE-INSENSITIVE1 (COI1), an F-box protein that is required for JA-signaled processes in Arabidopsis. These findings indicate that the JA/COI1 signaling pathway regulates distinct developmental processes in different plants and suggest a role for JA in the promotion of glandular trichome–based defenses.
INTRODUCTION
Plant oxylipins constitute a group of bioactive fatty acid derivatives that perform several important roles in growth and development. A large body of research has focused on the jasmonate family of oxylipins, which includes jasmonic acid (JA) and its methyl ester, methyl jasmonate (MeJA). These signaling compounds, collectively referred to as JAs, are ubiquitous in the plant kingdom and are well characterized with respect to their role in regulating defense responses against herbivore attack and infection by some pathogens (Kessler and Baldwin, 2002; Turner et al., 2002; Wasternack and Hause, 2002; Weber, 2002). JAs also are implicated in the control of plant responses to abiotic stimuli such as mechanical stress (Weiler et al., 1993), salt stress (Dombrowski, 2003), UV irradiation (Conconi et al., 1996), and ozone exposure (Rao et al., 2000). The ability of JAs to regulate gene expression in plant, insect (Li et al., 2002c), and mammalian (Rotem et al., 2003) cells indicates that some components of this lipid-based signaling system may be conserved in diverse biological systems.
Exogenous JAs exert numerous inductive and inhibitory effects on plant developmental processes (Creelman and Mullet, 1997; Wasternack and Hause, 2002). Correlations between endogenous JA levels in specific tissues and the effects of the applied hormone have provided evidence that JAs have a role in promoting senescence, fruit ripening, embryo development, and the accumulation of storage proteins (Staswick, 1990; Wilen et al., 1991; Creelman and Mullet, 1997; He et al., 2002). However, because exogenous JAs do not target specific cell types and often are administered at nonphysiological concentrations, confirmation of these roles requires genetic manipulation of either endogenous JA levels or the signal transduction steps that couple JA production to the physiological response.
Genetic analysis has been instrumental in elucidating the function of JA in Arabidopsis development. Initial insight into the role of JA in flower development came from the observation that the coronatine-insensitive1 (coi1) mutant that is insensitive to JAs is male sterile (Feys et al., 1994). The COI1 gene encodes an F-box protein that participates in the formation of an E3 ubiquitin ligase complex involved in ubiquitin-dependent proteolysis (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002). The critical requirement for JA in male fertility was established by the characterization of an Arabidopsis mutant that fails to produce linolenic acid, the fatty acid precursor of JA (McConn and Browse, 1996). Subsequently, mutations that disrupt other steps in the JA biosynthetic pathway also were shown to cause male sterility (Sanders et al., 2000; Stintzi and Browse, 2000; Ishiguro et al., 2001; Park et al., 2002). Reproductive dysfunction in JA-deficient Arabidopsis plants results from a combination of defects in anther filament elongation, anther dehiscence, and pollen maturation. Based on the temporal and spatial expression pattern of the DAD1 gene, which is required for JA biosynthesis and male fertility, it was proposed that JA-regulated water transport within floral tissues may promote synchronous pollen maturation, anther dehiscence, and flower opening (Ishiguro et al., 2001). The molecular mechanism by which JA promotes male reproductive development remains unknown. Recent studies support the hypothesis that JA controls the expression of genes that are required for normal anther development and pollen maturation (Mandaokar et al., 2003).
A more complete understanding of the physiological function of JA in plant growth and development would be facilitated by the identification of JA signaling mutants in diverse plant species. Toward this goal, we previously reported the identification of tomato mutants that are insensitive to JAs (Li et al., 2001). The recessive jasmonic acid–insensitive1-1 (jai1-1) mutant was isolated in a genetic screen for plants that fail to accumulate defense-related proteins in response to volatile MeJA. A second, allelic mutation (jai1-2) was identified as a suppressor of the systemin-mediated signaling pathway that requires JA biosynthesis and action to promote anti-herbivore defense responses in tomato (Howe and Ryan, 1999; Li et al., 2002b). In contrast to JA signaling mutants of Arabidopsis, reciprocal crosses showed that jai1 plants are male fertile and female sterile (Li et al., 2001). This observation suggested that JA has different roles in the reproductive development of different plant species. On the other hand, several defense-related phenotypes of jai1 plants are similar to those of Arabidopsis coi1 mutants, including loss of expression of defense-related genes in response to wounding and MeJA, insensitivity to the phytotoxin coronatine (COR), and increased resistance to virulent strains of Pseudomonas syringae (Li et al., 2001; Zhao et al., 2003).
Here, we report that jai1-1 and jai1-2 correspond to mutations in the tomato homolog of the Arabidopsis COI1 gene. These results demonstrate that this gene is essential for JA-regulated gene expression and anti-herbivore defense. We also show that tomato COI1 plays a role in the maternal control of seed maturation and the development of glandular trichomes. The potential involvement of JA in the regulation of these developmental processes is discussed.
RESULTS
jai1-1 Plants Are Insensitive to Exogenous Jasmonates
The jai1-1 mutant was identified in a screen of a fast-neutron-mutagenized population of Micro-Tom plants for individuals that are deficient in the accumulation of polyphenol oxidase (PPO) and the Ser proteinase inhibitor (PI), PI-II, in response to exogenous MeJA (see Methods) (Li et al., 2001). With the exception of the overt phenotypes described below, jai1-1 homozygotes exhibited normal vegetative morphology and growth (Figure 1A). F1 plants derived from a cross between jai1-1 pollen and a wild-type female parent accumulated normal levels of leaf PPO and PI-II in response to MeJA treatment. Semiquantitative analysis of leaf PPO activity in 687 adult F2 plants (Figure 1B) showed that the ratio of MeJA-responsive to MeJA-nonresponsive plants was 519:168 (χ2 = 0.12, P = 0.72 for the 3:1 hypothesis), indicating that jai1-1 is a single recessive mutation.
Figure 1.
jai1-1 Plants Are Insensitive to MeJA.
(A) Morphology of 4-week-old Micro-Tom (wild type [WT]; left) and jai1-1 (right) plants.
(B) Rapid assay for MeJA-induced PPO activity in tomato leaves. Leaf juice from individual leaflets of 15-day-old plants was expressed on a nitrocellulose membrane and assayed for PPO activity as described by Howe and Ryan (1999). Wild-type plants were treated with MeJA (+) or ethanol (−) as a control. F2 plants derived from a cross between jai1-1 and the wild type were treated with MeJA before testing for PPO activity. Dark brown staining indicates the presence of PPO activity.
(C) Response of germinating seedlings to MeJA. Wild-type, heterozygous (J/j), and homozygous (j/j) seeds were germinated on moist filter paper and exposed to MeJA (+) for 2 days. A wild-type seedling grown in the absence of MeJA (−) is shown as a control.
To determine whether jai1-1 affects seedling responses to JAs, we treated 4-day-old wild-type and jai1-1 F2 seedlings (n = 132) with a 1-mM solution of MeJA. All MeJA-treated wild-type seedlings showed inhibition of root elongation, reduced hypocotyl growth, and accumulation of anthocyanin in the hypocotyl (Figure 1C). Approximately three-fourths of the F2 population (104 seedlings) exhibited this MeJA-sensitive phenotype, whereas the remaining one-fourth (28 seedlings) were unaffected by the hormone (Figure 1C). After transfer to soil and growth for an additional 3 weeks, we found that MeJA-treated leaf tissue from each of the JA-insensitive plants lacked detectable PPO and PI-II accumulation (data not shown). These results demonstrate that jai1-1 disrupts multiple JA-signaled responses in seedlings and mature plants.
Reproductive Phenotypes of jai1-1 Plants
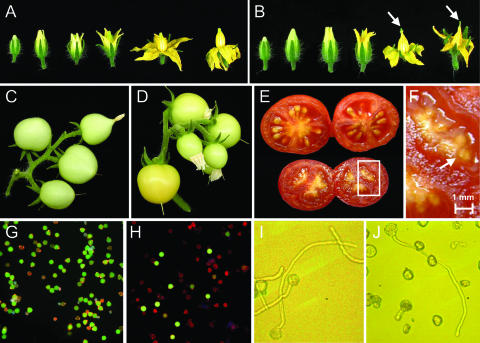
The general morphology and timing of development of jai1-1 flowers were very similar to those of wild-type flowers (Figures 2A and 2B). However, the tip of mature jai1-1 anther cones exhibited tissue collapse and browning, and the stigmas of mutant flowers protruded from the anther cone during later stages of flower development. Despite these anomalies, jai1 flowers readily initiated fruit development upon pollination. The size of immature jai1 fruit was similar to that of the wild type (Figures 2C and 2D), as was the general timing of fruit ripening. However, the size and mass of mature ripened jai1-1 fruit were significantly less than those of mature wild-type fruit (Figure 2E). In one set of measurements using plants grown under identical conditions, the average weight of ripe fruit from wild-type and jai1-1 plants was 5.4 ± 1.1 g and 2.9 ± 0.4 g, respectively (mean ± sd; n = 100 fruit/genotype; P < 0.0001). Wild-type (Micro-Tom) fruit yielded ∼30 seeds, each having a dry weight of ∼2.5 mg. Ripe jai1-1 fruit contained small, undeveloped seeds weighing <0.7 mg dry weight (Figure 2F). Although the vast majority (>99%) of these were not viable, a few viable seeds were recovered from jai1-1 fruit (Li et al., 2001) (data not shown). We estimated that the number of viable seeds produced by jai1-1 plants was <0.1% of the viable seed yield from wild-type plants grown under identical conditions, which for all practical purposes constitutes a sterile phenotype. Examination of several hundred fruit-bearing jai1-1 homozygotes obtained from three successive backcrosses showed that the sterile phenotype strictly cosegregated with the JA-insensitive phenotypes in leaves (data not shown).
Figure 2.
Reproductive Phenotypes of jai1-1 Plants.
(A) and (B) Developmental progression of wild-type (A) and jai1-1 (B) flowers. Arrows in (B) show the stigma protruding from the anther cone.
(C) and (D) Developing fruit on wild-type (C) and jai1-1 (D) plants.
(E) Mature wild-type (top) and jai1-1 (bottom) fruit.
(F) Enlargement of jai1-1 fruit in (E), showing small undeveloped seeds.
(G) and (H) Fluorescein diacetate/propidium iodide costaining showing viable (green) and nonviable (red) pollen from wild-type (G) and jai1-1 (H) anthers.
(I) and (J) Light microscopic images (×100 magnification) of germinated wild-type (I) and jai1-1 (J) pollen.
Reciprocal crosses between wild-type and jai1-1 flowers showed that the mutation disrupts the maternal control of seed maturation (Li et al., 2001). We considered the possibility that this defect might be caused by an interaction between jai1 and one of the mutations responsible for the dwarf stature of Micro-Tom (Meissner et al., 1997). However, after four successive backcrosses of jai1-1 pollen to the standard Castlemart cultivar, jai1-1 homozygotes selected from a resulting F2 population remained sterile (data not shown). Thus, we conclude that jai1-1 is sufficient to disrupt the maternal control of seed development.
The expression of PI-I and PI-II in tomato and potato leaves is induced in response to wounding and JA but is constitutive in flowers and tubers (Ryan, 1973; Peña-Cortés et al., 1991). The absence of PI-II accumulation in MeJA-treated jai1-1 leaves suggested that the mutant also might be deficient in the constitutive expression of this protein in flowers. Using a radial immunodiffusion assay, we determined that the PI-II content of newly opened flower buds from wild-type plants was 218 ± 52 μg/mL total soluble protein (n = 15 flowers). By contrast, PI-II accumulation in similarly staged jai1-1 flowers was undetectable. This finding indicates that jai1-1 abrogates constitutive PI-II expression in flowers, which is correlated with female sterility.
Because Arabidopsis mutants that are impaired in JA synthesis or JA perception are male sterile (Feys et al., 1994; McConn and Browse, 1996), we examined jai1-1 plants for possible defects in pollen development. Staining of pollen grains with Alexander's triple stain showed that the proportion of nonaborted pollen grains (i.e., containing cytoplasm) from similarly staged wild-type and jai1-1 anthers was 94 and 67%, respectively (data not shown). Fluorescein diacetate/propidium iodide costaining further showed a significant reduction (P < 0.005) in the viability of jai1-1 pollen (28% viability) compared with wild-type pollen (82% viability; Figures 2G and 2H). The general trend toward reduced vigor of jai1-1 pollen was reflected in measurements of in vitro germination rates. In three independent experiments, the germination rate of jai1-1 was ∼10%, whereas that of wild-type pollen was >55%. The morphology and tube length of germinated jai1-1 pollen grains were similar to those of wild-type pollen (Figures 2I and 2J). Together with the results from reciprocal cross experiments (Li et al., 2001), these findings indicate that viable pollen production in jai1-1 plants is reduced but nevertheless sufficient for male reproductive function.
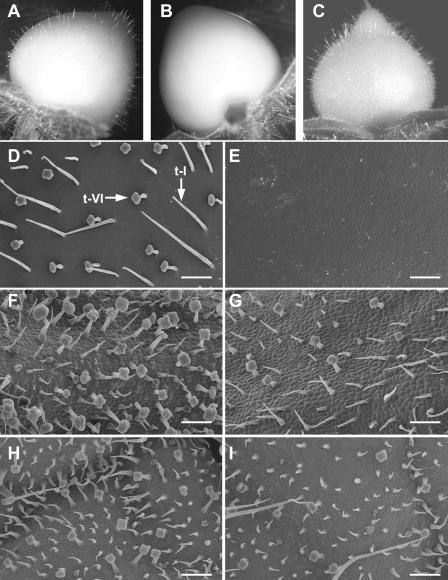
jai1-1 Affects the Development of Glandular Trichomes
Epidermal cells of immature tomato fruit give rise to various types of trichomes, which disappear upon fruit maturation. We observed two morphologically distinct multicellular trichome types on immature (green) wild-type fruit (Figures 3A and 3D): the hair-like glandular type-I trichome consisting of an elongated multicellular stalk and a small unicellular vesicle at the tip, and the well-characterized type-VI glandular trichome containing a short stalk and a four-celled glandular head (Luckwill, 1943; Kennedy, 2002). A striking feature of developing jai1-1 fruit was the complete absence of both trichome types, resulting in a smooth and glabrous appearance (Figures 3B and 3E). We also observed a significant reduction in the density of type-VI trichomes on jai1-1 leaves and sepals (Figures 3F to 3I). Quantification of trichome density on leaflets of 6-week-old wild-type plants (containing at least seven leaves) showed that densities ranged from ∼35 type-VI trichomes/mm2 on terminal leaflets of young leaves to ∼3 type-VI trichomes/mm2 on leaflets from older leaves. Analysis of comparable jai1-1 leaflets showed that, irrespective of leaf age, type-VI trichome density on the mutant was 25 to 35% of that of wild-type leaflets. Similarly, the density of type-VI trichomes on wild-type and jai1 sepals of the same developmental stage was 20 ± 3 and 7 ± 3 per mm2, respectively (P < 0.001).
Figure 3.
jai1-1 Plants Exhibit Defects in Trichome Development.
(A) to (C) Photographs of developing green fruit from wild-type (A), jai1-1 (B), and 35S-LeCoi1–complemented jai1-1 (C) plants.
(D) and (E) Scanning electron micrographs of the surface of developing green fruit from wild-type (D) and jai1-1 (E) plants. Arrows in (D) denote type-VI (t-VI) and type-I (t-I) trichomes. The tissue fixation procedure used for scanning electron microscopy affected the structure of type-I trichomes, which were 1 to 2.5 mm long on intact tissue (see [A] and [D]).
(F) and (G) Scanning electron micrographs of wild-type (F) and jai1-1 (G) sepals.
(H) and (I) Scanning electron micrographs of the adaxial (upper) side of young leaves from wild-type (H) and jai1-1 (I) plants.
Bars in (D) to (I) = 0.2 mm.
To determine whether jai1-1 affects the production of compounds that are synthesized in trichome glands, we used gas chromatography to measure the content of volatile terpenes in exudates obtained by brief extraction of tissue with tert-methyl butyl ether (MTBE). Six compounds were detected in exudates from wild-type fruit. Four of these were identified as the monoterpenes α- and β-pinene, limonene, and cis-β-ocimene (Figure 4A), which were shown previously to be volatile components of tomato leaf aroma (Andersson et al., 1980; Buttery et al., 1987). Analysis of extracts obtained either from wiped wild-type fruit (to remove trichomes) or directly from type-VI glands demonstrated that the monoterpenes were derived from glandular trichomes (Figures 4E and 4F). Monoterpene accumulation was not detected in extracts from jai1 fruit (Figure 4B), consistent with the lack of trichomes on this tissue. The monoterpene content of trichome exudates from wild-type sepals was comparable to that of developing fruit (Figure 4C). By contrast, the monoterpene level in jai1 sepals was reduced significantly compared with that in wild-type sepals (Figure 4D).
Figure 4.
Monoterpene Content in Trichome Exudates.
Trichome exudates obtained by brief extraction of tissues with MTBE were analyzed for monoterpene content by gas chromatography–mass spectrometry.
(A) Wild-type (WT) green fruit.
(B) jai1-1 green fruit.
(C) Wild-type sepal.
(D) jai1-1 sepal.
(E) Before extraction with MTBE, wild-type fruit was wiped with a cotton swab to remove trichome contents.
(F) Exudate collected directly from type-VI trichome glands of wild-type green fruit. The y axis indicates arbitrary absorbance units (AU).
jai1-1 Plants Are Defective in JA-Induced Gene Expression
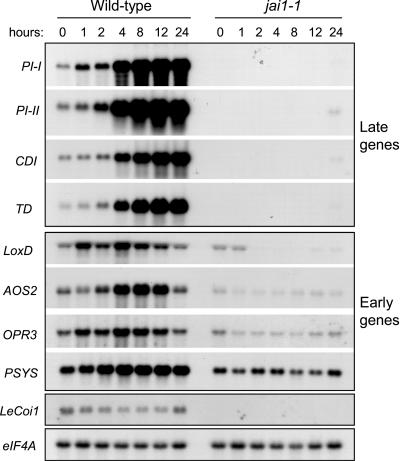
RNA gel blot analysis was used to determine the effect of jai1-1 on the MeJA-induced expression of several defense-related genes that are induced either “early” or “late” in response to wounding of tomato leaves (Ryan, 2000; Lee and Howe, 2003). Late gene transcripts, including PI-I, PI-II, CATHEPSIN D INHIBITOR, and THREONINE DEAMINASE, began to accumulate within 2 to 4 h of MeJA treatment and reached maximum levels 12 to 24 h after treatment. With the exception of very low levels of mRNA accumulation (<2% of wild type) at the 24-h time point, the expression of late genes in MeJA-treated and untreated control jai1-1 plants was undetectable. Treatment of wild-type plants with MeJA caused moderate and transient expression of early wound response genes, including those that encode the octadecanoid pathway enzymes LIPOXYGENASE D (LoxD; Heitz et al., 1997), ALLENE OXIDE SYNTHASE2 (AOS2; Howe et al., 2000), 12-OXO-PHYTODIENOIC ACID REDUCTASE3 (OPR3; Strassner et al., 2002), and PROSYSTEMIN (PSYS; Jacinto et al., 1997). In jai1-1 plants, MeJA treatment either reduced (e.g., LoxD) or did not significantly affect (e.g., PSYS) the accumulation of early gene transcripts (Figure 5). It also was apparent that, unlike the late genes, jai1-1 plants maintained a basal level of expression of early response genes.
Figure 5.
MeJA-Induced Gene Expression in Wild-Type and jai1-1 Plants.
Three-week-old Micro-Tom and jai1-1 plants were exposed to MeJA vapor for various lengths of time (hours) in an enclosed box. Leaves from similarly treated plants of the same genotype were pooled for RNA extraction. RNA isolated from untreated plants (0-h time point) also was analyzed as a control. RNA gel blots were hybridized to cDNA probes representing four late genes (PI-I, PI-II, CATHEPSIN D INHIBITOR [CDI], and THREONINE DEAMINASE [TD]) and four early genes (LIPOXYGENASE D [LoxD], ALLENE OXIDE SYNTHASE2 [AOS2], OPDA REDUCTASE3 [OPR3], and PROSYSTEMIN [PSYS]). Blots also were hybridized to the LeCoi1 cDNA and, as a loading control, to an eIF4A probe.
DNA microarray analysis was used to further examine the effect of jai1-1 on MeJA-induced gene expression. These experiments used a microarray slide containing 607 tomato cDNAs corresponding to ∼500 unique genes involved in various aspects of herbivore and pathogen defense, signal transduction, lipid metabolism, and hormone synthesis (Zhao et al., 2003). A complete list of these genes together with the expression data for each is provided in the supplemental data online. To identify jasmonate-responsive genes among this collection of sequences, slides were hybridized simultaneously to probes that were prepared from plants treated for 8 h with MeJA or a mock control (ethanol). This relatively brief exposure time was chosen to minimize possible secondary effects resulting from MeJA application and because both early and late genes are upregulated at this time point (Figure 5). Analysis of data from two independent biological replicates showed that 37 genes were upregulated >2.5-fold in MeJA-treated wild-type plants (Table 1; see also supplemental data online). This list included the four early genes and four late genes analyzed by RNA gel blot analysis (Figure 5) as well as other previously identified JA- and wound-responsive genes, including LEUCINE AMINOPEPTIDASE (LAP; Chao et al., 1999), METALLOCARBOXYPEPTIDASE INHIBITOR (MCPI; Villanueva et al., 1998), ALLENE OXIDE CYCLASE (AOC; Hause et al., 2000), LIPOXYGENASE A (LOXA; Beaudoin and Rothstein, 1997), POLYPHENOL OXIDASE-F (PPO-F; Constabel et al., 1995), a wound-inducible SERINE CARBOXYPEPTIDASE (CP; Moura et al., 2001), CYSTATIN (Schaller et al., 1995), and AOS1 (Sivasankar et al., 2000). Remarkably, none of these 37 genes was upregulated differentially in MeJA-treated jai1-1 plants (Table 1; see also supplemental data online). This finding indicates that the gene defined by jai1 plays a major role in promoting the expression of the JA-regulated transcriptome in tomato.
Table 1.
Microarray Analysis of Genes Induced by MeJA in Wild-Type and jai1-1 Plants
| Expression Ratio
|
|||
|---|---|---|---|
| Accession No. | Gene Product (Best BLAST Hit) | Wild Type | jai1 |
| AI485116 | Thr deaminase | 41.9 | – a |
| Q10712 | Leu aminopeptidase A (LAP-A) | 33.0 | 1.3 |
| AI485529 | Putative acyltransferase | 25.9 | 0.7 |
| AI486173 | Protein translation inhibitor | 19.8 | – a |
| AI897750 | Kunitz-type trypsin inhibitor | 14.8 | 0.8 |
| AW037833 | Metallocarboxypeptidase inhibitor | 12.7 | – a |
| AI487422 | Pto-responsive gene 1 protein | 12.5 | 1.0 |
| AI488657 | Cathepsin D inhibitor protein (CDI) | 12.4 | 0.7 |
| K03291 | Proteinase inhibitor II (PI-II) | 11.2 | – a |
| AW649914 | Leu aminopeptidase N (LAP-N) | 11.0 | 1.3 |
| AW624058 | Allene oxide cylase (AOC) | 10.8 | 1.4 |
| U09026 | Lipoxygenase A (LOXA) | 9.2 | 0.8 |
| Z12838 | Polyphenol oxidase F (PPO-F) | 8.4 | 1.3 |
| K03290 | Proteinase inhibitor (PI-I) | 8.2 | 1.0 |
| AW092579 | Nucleoside diphosphate kinase | 5.7 | 1.0 |
| AI897184 | Glyoxylase family protein | 5.5 | 0.9 |
| AI490318 | NAC domain protein | 5.5 | 0.8 |
| AI486025 | 4-Coumarate:CoA ligase | 5.2 | 1.1 |
| AW040669 | Thioredoxin M-type 3 chloroplast precursor |
5.1 | 0.9 |
| AI486916 | Kunitz-type enzyme inhibitor | 4.5 | 1.1 |
| AI486546 | Wound-inducible carboxypeptidase (WIC) |
4.5 | 1.1 |
| AF198389 | Cystatin | 4.4 | 1.1 |
| AI489221 | Wound-inducible WRKY transcription factor |
4.3 | 1.0 |
| AI483527 | Kunitz-type trypsin inhibitor | 4.2 | 0.7 |
| Z21793 | DAHP-synthase 2 | 4.2 | 1.2 |
| AW034958 | 12-Oxo-phytodienoate reductase3 (OPR3) |
4.2 | 1.1 |
| AI771886 | RD2 auxin-regulated protein | 4.0 | 1.1 |
| AW032472 | Unknown protein, similar to PnFL-2 |
4.0 | 0.9 |
| AW220064 | Glutathione S-transferase | 3.9 | 1.2 |
| AF230371 | Allene oxide synthase2 (AOS2) | 3.8 | 1.0 |
| AW648326 | Adenosylmethionine decarboxylase |
3.5 | 1.1 |
| U37840 | Lipoxygenase D (LOXD) | 3.4 | 1.0 |
| AI895589 | Allene oxide synthase1 (AOS1) | 3.0 | 0.9 |
| M84800 | Prosystemin | 2.9 | 1.2 |
| AI897620 | Putative chorismate mutase | 2.9 | 1.2 |
| AI483536 | TMV response-related gene product |
2.7 | 0.9 |
| BE459901 | Putative caffeoyl-CoA O-methyltransferase |
2.6 | 0.7 |
Three-week-old Castlemart (wild-type) and jai1-1 plants were treated with either MeJA or ethanol (mock control) for 8 h. Leaf tissue was harvested for RNA isolation after the treatment. A custom cDNA microarray slide representing ∼500 tomato genes was hybridized simultaneously to probes derived from RNA isolated from ethanol- and MeJA-treated plants of the same genotype (i.e., wild type or jai1-1). Numbers represent the mean expression ratio (MeJA:ethanol) of two independent biological replicates for each experiment. Genes (GenBank accession number and putative function) that were differentially regulated by >2.5-fold in response to MeJA in wild-type plants are listed.
Expression was not detectable, indicating that the microarray signal was below background levels.
jai1-1 Causes Susceptibility to Spider Mites and Modulates Mite Behavior
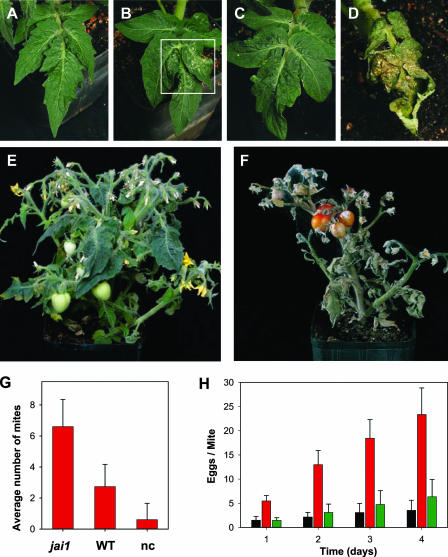
The lack of MeJA-induced gene expression in jai1-1 plants suggested that the mutant might be compromised in resistance to herbivore attack. To test this idea, we used various biological assays to compare the interaction of wild-type and jai1 genotypes with the two-spotted spider mite, a cell-content-feeding arachnid that is a major pest of many crop plants. Two days after challenge with adult female mites, small (<0.5 mm) chlorotic lesions indicative of mite feeding were evident on jai1-1 but not on wild-type leaves (Figures 6A and 6B). Localized collapse of jai1-1 tissue was apparent 6 days after infestation, with total collapse and desiccation of the leaf occurring by 11 days (Figures 6C and 6D). Longer-term feeding trials, in which spider mites dispersed to aerial tissues, resulted in severe damage to and eventual death of jai1-1 plants (Figure 6F). Wild-type plants showed relatively few signs of macroscopic damage during the course of these feeding trials (Figures 6C and 6E). These results indicate that Micro-Tom plants have a relatively high level of natural resistance to the two-spotted spider mite and that this resistance is severely compromised by jai1-1.
Figure 6.
Resistance of jai1-1 Plants to the Two-Spotted Spider Mite Is Severely Compromised.
(A) to (D) Fifteen adult female mites were transferred to a single leaf on 3-week-old wild-type ([A] and [C]) and jai1-1 ([B] and [D]) plants. Photographs of the infested leaves were taken 6 days ([A] and [B]) and 11 days ([C] and [D]) after challenge. The boxed area in (B) shows the initial effects of feeding damage on jai1-1.
(E) and (F) Photographs of representative 7-week-old wild-type (E) and jai1-1 (F) plants at 30 days after infestation.
(G) Two-choice assay to measure the preference of spider mites for wild-type (WT) or jai1-1 plants. Ten adult female spider mites were placed in an arena equidistant from wild-type and jai1-1 leaflets. The number of mites that moved to one or the other of the leaflets was determined 1 h after initiating the assay, as was the number of mites that failed to make a choice (nc). Data represent means and standard errors from 16 repetitions (total of 160 mites).
(H) Fecundity of two-spotted spider mites on wild-type (black bar), jai1-1 (red bar), and 35S-LeCoi1–complemented jai1-1 (green bar) plants. Data represent means and standard deviations from 12 experimental repetitions in which five teneral female mites were reared for 4 days on leaf discs of the indicated host genotype.
To further characterize the effect of jai1-1 on spider mite performance, we used a two-choice assay to assess the relative preference of the herbivore for wild-type and jai1-1 leaves. Within 1 h of initiating the assay, >94% of the mites were found either on or under one of the two leaflets. However, the mites selected jai1 over wild-type leaves in 99 of 140 plant visits (Figure 6G) (P < 0.05). Even distributions of mites were observed when leaflet pairs of the same genotype were used, with no significant difference in the number of visitations to either genotype (data not shown). This observation indicates that neither genotype is particularly repellent to mites, but rather that jai1 plants are more attractive than are wild-type plants. The ability of the herbivore to colonize the host was examined by measuring the fecundity of female mites on wild-type and jai1 leaves. The results showed that the number of eggs laid on jai1 leaves during the 4-day time course was significantly greater than that on wild-type leaves (Figure 6H).
jai1 Disrupts the Function of the Tomato Homolog of COI1
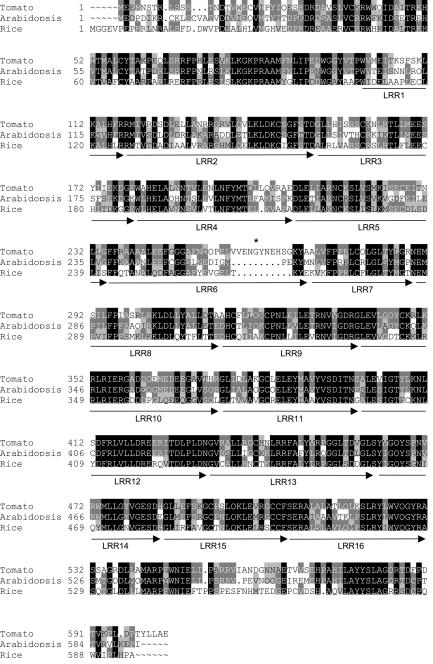
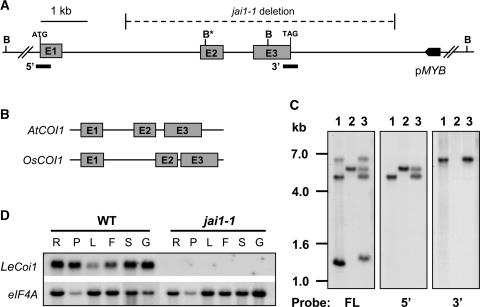
The effect of jai1-1 on MeJA-induced gene expression, resistance to herbivores, and responses to COR (Zhao et al., 2003) suggested that the mutant might be defective in the tomato homolog of the COI1 protein that is required for JA-signaled processes in Arabidopsis (Xie et al., 1998). To test this possibility, we isolated a cDNA that encodes the tomato homolog, designated LeCOI1 (Lycopersicon esculentum COI1; see Methods). The full-length cDNA contained a 1812-nucleotide open reading frame predicted to encode a 603–amino acid protein. A BLAST (Basic Local Alignment Search Tool) search (Altschul et al., 1990) of this sequence against the NCBI nonredundant protein database identified unambiguous matches (E value = 0) to Arabidopsis COI1 and a putative homolog predicted from the rice genome sequence. Amino acid sequence alignments showed that LeCOI1 is 68 and 57% identical to the Arabidopsis and rice proteins, respectively (Figure 7). Similar to Arabidopsis COI1 (Xie et al., 1998), the predicted COI1 proteins in tomato and rice contain an N-terminal F-box domain and 16 imperfect Leu-rich repeats (LRRs). To obtain additional evidence for homology between tomato and Arabidopsis COI1, we determined the DNA sequence of the tomato gene (LeCoi1) that was identified on a BAC clone. Comparison of the genomic and cDNA sequences showed that LeCoi1 comprises three exons and two introns that span ∼6 kb (Figure 8A). The positions of the two introns within the gene, together with the coding capacity of individual exons, were identical to those of the Arabidopsis and rice genes (Figure 8B). Intron size accounted for the major differences between the three genes; on average, the size of introns in LeCoi1 was threefold greater than the corresponding introns in the Arabidopsis and rice genes (Figure 8B).
Figure 7.
Deduced Amino Acid Sequence of LeCOI1 Compared with Homologs in Arabidopsis and Rice.
Sequence alignments were performed using the GCG sequence analysis package (Genetics Computer Group, Madison, WI). Amino acids that are either identical or similar between the three sequences are indicated in black and gray, respectively. Arrows denote the approximate positions of the 16 imperfect LRR domains. The arrow and LRR domain number are indicated beneath the corresponding LRR. The asterisk denotes the Gly residue that is changed to a Cys in jai1-2 plants. GenBank accession numbers are given at the end of Methods. The N-terminal end of the rice sequence contains 35 additional amino acids that are not included in the alignment.
Figure 8.
The jai1-1 Mutation Is a 6.2-kb Deletion in LeCoi1.
(A) Scheme of a 9.4-kb region of genomic DNA containing LeCoi1 and a gene encoding a putative MYB transcription factor (pMYB). The three exons (E1 to E3) and intervening sequences that constitute LeCoi1 are drawn to scale. Only the translated portion of the first and last exons are shown, together with the start (ATG) and stop (TAG) codons within the respective exons. The 6.2-kb region of DNA that is deleted in jai1-1 plants is shown by the horizontal hatched line. B indicates BglII restriction sites used for the DNA gel blot analysis shown in (C), and B* indicates the presence of two BglII sites that are separated by 24 bp. cDNA probes used to detect the 5′ and 3′ ends of the gene in (C) are shown as thick horizontal bars.
(B) Structures of the Arabidopsis (AtCOI1) and rice (OsCOI1) homologous genes drawn to the same scale as LeCoi1 in (A).
(C) DNA gel blot analysis of BglII-restricted genomic DNA from the wild type (lanes 1), jai1-1 (lanes 2), and an F1 hybrid produced from a cross between the wild type and jai1-1 (lanes 3). Three identical blots were hybridized to a full-length (FL) LeCoi1 cDNA probe or to probes corresponding to the 5′ and 3′ ends of the cDNA (see [A]). Numbers at left indicate the positions of DNA size standards (in kb).
(D) RNA gel blot analysis of LeCoi1 transcript levels in wild-type (WT) and jai1-1 plants. Five micrograms of total RNA from root (R), petiole (P), leaf (L), unopened flower bud (F), sepal (S), and immature green fruit (G) was immobilized to a membrane and hybridized to a LeCoi1 cDNA probe (top gel). A duplicate blot was hybridized to an eIF4A cDNA probe as a loading control.
Genomic DNA gel blot analysis was used to determine whether the fast-neutron bombardment used to create jai1-1 plants caused a deletion mutation at the LeCoi1 locus. The full-length LeCoi1 cDNA hybridized to three BglII restriction fragments (∼6.5, 4.5, and 1.3 kb) in wild-type genomic DNA but only a single 5.5-kb fragment in DNA from a jai1-1 homozygote (Figure 8C). Hybridization experiments using cDNA end–derived probes further indicated that genomic DNA from jai1-1 plants contains the 5′ end but not the 3′ end of LeCoi1 (Figure 8C). To define the size of this deletion polymorphism more precisely, thermal asymmetric interlaced PCR was used to isolate a fragment of genomic DNA from jai1-1 plants that spans the deletion end points. Comparison of the sequence of a resulting 1.3-kb PCR product with the LeCoi1 genomic sequence showed that the deletion spans 6243 bp and includes exons 2 and 3 (Figure 8A). The 3′ end of the deletion was mapped 2.3 kb downstream of the LeCoi1 stop codon. Sequence analysis using BLAST searches indicated that the deletion does not affect genes other than LeCoi1, including a putative MYB factor gene located ∼0.8 kb downstream of the deletion end point. RNA gel blot analysis demonstrated that LeCoi1 transcripts, which accumulate in all wild-type tissues examined, do not accumulate in jai1-1 plants (Figures 5 and 8D).
To determine whether the ethyl methanesulfonate–induced jai1-2 mutation corresponds to a lesion in LeCoi1, we used reverse transcription–PCR to obtain full-length cDNAs from jai1-2 homozygotes. DNA sequencing of these clones revealed a single G-to-T transversion that results in the replacement of Gly-261, which is located between the sixth and seventh LRRs, by a Cys (Figure 7). This polymorphism was verified by sequencing the corresponding genomic region from wild-type and jai1-2 plants.
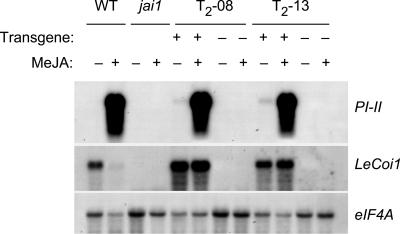
To confirm that jai1-associated phenotypes result from the loss of function of LeCOI1, we used Agrobacterium tumefaciens to transform jai1-1 plants with the LeCoi1 cDNA under the control of the 35S promoter of Cauliflower mosaic virus. From 28 independent 35S-LeCoi1 transgenic plants confirmed by genomic PCR, nine primary lines (T1) exhibited trichome development on immature fruit (Figure 3C) and normal seed production. Gas chromatographic analysis of trichome exudates from three of these lines showed that monoterpene accumulation was restored (data not shown). Additional phenotypic characterization was performed on T2 progeny derived from two primary lines (T2-08 and T2-13), which by DNA gel blot analysis were shown to harbor one or two copies of the transgene (data not shown). RNA gel blot experiments demonstrated that T2 plants expressing 35S-LeCoi1 accumulated PI-II mRNA in response to exogenous MeJA, whereas T2 siblings lacking the transgene did not (Figure 9). The results of this and other (Figure 5) RNA gel blot hybridization experiments also showed that exogenous MeJA reduced the abundance of LeCoi1 transcripts in wild-type plants. This effect was not observed in the transgenic lines in which LeCoi1 was expressed from the 35S promoter. Consistent with the restoration of JA-induced PI expression in 35S-LeCoi1–complemented lines, we also found that the fecundity of female spider mites on these lines was reduced to the level observed on wild-type plants (Figure 6H). These genetic complementation studies demonstrate that Jai1 and LeCoi1 are equivalent and that this gene promotes multiple processes in tomato, including sensitivity to JAs, resistance to herbivores, glandular trichome development, and seed maturation.
Figure 9.
MeJA-Induced Gene Expression in 35S-Coi1–Complemented Transgenic Lines.
Total RNA was prepared from wild-type (WT), jai1-1 (jai1), and T2 siblings from two 35S-Coi1–complemented lines (T2-08 and T2-13) either before (−) or after (+) exposure to MeJA vapor for 12 h. For each transgenic line tested, a PCR assay was used to identify siblings that either harbor (+) or lack (−) the 35S-LeCoi1 transgene. RNA gel blots were hybridized to cDNA probes for PI-II and LeCoi1 and to eIF4A as a loading control.
DISCUSSION
Identification of a Tomato Homolog of COI1
We report here the phenotypic and molecular characterization of the tomato jai1 mutant that was identified in a genetic screen for mutants that fail to produce PPO and PI-II in response to MeJA. Using a candidate gene approach that relied on previous knowledge of JA signaling in Arabidopsis, we demonstrate that jai1 disrupts the function of a tomato homolog (LeCOI1) of COI1. The role of Jai1/LeCoi1 (henceforth referred to as LeCoi1) in promoting the responsiveness of roots to JA, JA-induced gene expression in leaves, and the accumulation of JA-regulated PIs in flowers indicates that this gene mediates JA-signaled responses in most, if not all, tissues of tomato. This conclusion is supported by the accumulation of LeCoi1 transcripts in all major organs of wild-type plants (Figure 8D).
Recent studies with Arabidopsis have established that COI1 is part of a multiprotein complex that functions as an E3-type ubiquitin ligase (Xie et al., 1998; Devoto et al., 2002; Xu et al., 2002). This assembly of proteins, referred to as SCFCOI (for Skp1, Cullin, and F-box protein) is proposed to work together with ubiquitin-activating (E1) and ubiquitin-conjugating (E2) enzymes to attach ubiquitin to a protein substrate that is recruited to the complex via interaction with the LRR domain of COI1 and subsequently degraded by the 26S proteosome (Creelman and Rao, 2002; Turner et al., 2002). Ubiquitin-dependent proteolysis plays an important role in modulating the abundance of regulatory proteins and recently emerged as a major theme in many aspects of plant signaling (Hare et al., 2003). Based on the strong JA-insensitive phenotypes of tomato jai1 and Arabidopsis coi1 mutants, we conclude that COI1 performs a similar function in JA signal transduction in these two species. The existence of a COI1 homolog in rice further indicates that this signaling pathway likely is operational in monocotyledonous plants.
The identification of jai1-1 as a 6.2-kb deletion in LeCoi1 is consistent with the origin of this allele from a fast-neutron-mutagenized population. Genetic complementation of jai1-1 with a 35S-LeCoi1 transgene demonstrated that the loss of function of COI1 accounts for the defense-related and developmental phenotypes of the mutant. This conclusion also is supported by the existence of a second jai1 mutation, the ethyl methanesulfonate–induced jai1-2 allele, which also disrupts JA signaling. It is significant that jai1-2 was identified on the basis of its ability to suppress the expression of PPO and PIs in response to systemin and its precursor protein, prosystemin (Howe and Ryan, 1999). Given the results reported here, we conclude that LeCOI1 is essential for promoting the systemin-mediated activation of defense genes. This is in agreement with other biochemical and genetic evidence showing that systemin action requires JA (Ryan, 2000; Lee and Howe, 2003; Li et al., 2003). The Gly-to-Cys amino acid substitution caused by jai1-2 is located in a region of the LRR domain that was shown previously to be important for JA signaling in Arabidopsis as well as for interaction between COI1 and putative SCFCOI1 substrates (Devoto et al., 2002; Ellis and Turner, 2002). Notably, however, this substitution is located within an eight–amino acid segment that is missing from both the Arabidopsis and rice sequences (Figure 7). Because the mutation causes the loss of JA responsiveness, it is possible that this region of LeCOI1 plays a role in recruiting protein substrates for ubiquitination. An alternative explanation is that the Cys created by jai1-2 forms a disulfide linkage that disrupts the structure and function of the protein.
COI1-Regulated Gene Expression
PIs and PPOs constitute an important anti-herbivore defense system in tomato (Duffey, 1986; Ryan, 2000). The inability of jai1 plants to express PIs and PPO in response to wounding or JA demonstrates that LeCOI1 is essential for this line of chemical defense. Interestingly, genes that encode PPO and PI-II are not present in the Arabidopsis genome (Allen, 2002; Van der Hoeven et al., 2002). This observation indicates that although components of the JA signaling pathway (i.e., COI1) are conserved in different plant species, many of the target genes regulated by the pathway are specific to a particular plant species or family. This idea is supported by numerous examples of JA-regulated phytochemical defense systems that operate in specific plant lineages (Heil et al., 2001; Memelink et al., 2001; Kessler and Baldwin, 2002; Wasternack and Hause, 2002; Goossens et al., 2003). Additional insight into the role of LeCOI1 in regulating the JA transcriptome in tomato was obtained by cDNA microarray analysis. These results showed that the MeJA-induced expression of each of the 37 genes identified in wild-type plants was dependent on LeCOI1 (Table 1), which demonstrates a central role for this F-box protein in the expression of JA-responsive genes in tomato. It should be noted that we identified two genes (encoding putative glucosyltransferases) that were induced by MeJA in jai1-1 plants (see supplemental data online). However, these genes were not regulated differentially in MeJA-treated wild-type plants and thus were not considered to be COI1-dependent genes.
Although our results demonstrate that LeCOI1 is essential for JA-induced gene expression in tomato, RNA gel blot analysis showed that the JA/COI1 signaling pathway performs different roles in controlling the expression level of early and late wound response genes. For example, both the basal and JA-induced expression of late genes (e.g., PIs) was abrogated in jai1-1 plants (Figure 5). By contrast, significant basal expression of early genes was maintained in jai1 plants. Furthermore, the expression of some of these genes (e.g., LoxD) decreased in jai1 plants in response to MeJA (Figure 5). This phenomenon may reflect the existence of a compensatory COI1-independent mechanism to restrict the expression of some wound/JA-responsive genes. The transient nature of the increase in early gene expression in MeJA-treated wild-type plants supports this view. The downregulation of LeCoi1 transcript levels in MeJA-treated wild-type plants (Figures 5 and 9) provides additional evidence that control points within the JA signaling pathway may be negatively regulated by JA.
Because jai1-1 is a null mutation in LeCoi1, we conclude that the expression of early genes is controlled by both COI1-dependent and COI1-independent mechanisms. This conclusion is consistent with previous studies showing that PSYS (an early gene) is expressed constitutively in unwounded tomato leaves and induced further in response to wounding and MeJA treatment (Jacinto et al., 1997). Similarly, JA biosynthetic enzymes, which also are encoded by early wound response genes, accumulate constitutively in unwounded tomato leaves (Stenzel et al., 2003). The COI1-independent basal expression of early genes suggests that the respective gene products serve an important function in the absence of stress conditions that trigger JA signaling. For example, uncoupling of the basal expression level of JA biosynthetic enzymes and prosystemin from the JA/COI1 pathway might provide a mechanism to ensure that the amplitude and timing of JA biosynthesis in response to herbivore attack is sufficient to activate downstream target genes. This hypothesis is consistent with evidence indicating that wound-induced JA synthesis in tomato and tobacco does not depend on the induced expression of JA biosynthetic genes (Miersch and Wasternack, 2000; Ziegler et al., 2001).
The critical role of LeCOI1 in JA-regulated gene expression is relevant to the observation that jai1-1 plants are insensitive to COR and highly resistant to COR-producing strains of P. syringae that cause bacterial speck disease on tomato (Zhao et al., 2003). Because these phenotypes of jai1-1 are essentially identical to those of Arabidopsis coi1 mutants (Feys et al., 1994; Kloek et al., 2001), the mechanism by which COI1 promotes responsiveness to COR and susceptibility to P. syringae likely is conserved between tomato and Arabidopsis. Studies conducted with Arabidopsis have shown that COR acts both to inhibit salicylic acid–mediated host defense responses and to enhance the formation of lesions (Kloek et al., 2001). Recent work performed with tomato supports this conclusion and further indicates that COR, acting as a JA mimic, promotes host susceptibility by specifically targeting the JA/COI1 signaling pathway (Zhao et al., 2003). In support of this view, comparison of the microarray results reported here with those presented by Zhao et al. (2003) shows that 36 of the 37 JA/COI1-responsive genes (Table 1) were induced in a COI1-dependent manner during infection of wild-type plants with a virulent strain of P. syringae. The similar pattern of COI1-dependent gene expression in MeJA-treated and P. syringae–infected tomato leaves is consistent with the observation that exogenous MeJA complements the loss of virulence of a COR-deficient strain of P. syringae (Zhao et al., 2003) and the idea that COR functions as a molecular mimic of JAs.
Role of COI1 in Tomato Reproduction
jai1 plants fail to produce viable seeds as a result of a defect in female reproductive development (Li et al., 2001). Functional complementation of the sterile phenotype with 35S-LeCoi1 indicates that COI1 is essential for this aspect of tomato reproductive function. The production of small aborted seeds in jai1 fruit (Figure 2F), together with the ability of jai1 ovules to be fertilized (B. McCaig and G. Howe, unpublished results), suggest that sterility results from arrest in embryo/seed maturation. Because cell division and expansion in fruit tissues is dependent on normal embryo and seed development (Gillaspy et al., 1993; Giovannoni, 2001), this hypothesis would explain the reduced size of jai1 fruit. A role for LeCOI1 in embryogenesis also is consistent with the abundance of JAs in tomato ovaries (Hause et al., 2000) as well as with previous studies implicating JA as an endogenous regulator of embryo development in oilseeds (Wilen et al., 1991; Hays et al., 1999). Whether endogenous JAs regulate reproductive function in tomato remains to be established. The fact that JA biosynthetic mutants of tomato do not exhibit female (or male) sterility does not disprove this hypothesis, because such mutants produce significant levels of JA in floral tissues (Li et al., 2003). Identification of tomato mutants that completely lack JA will be necessary to address this question. On the other hand, the disruption of either JA biosynthesis or COI1 function in Arabidopsis does not affect female reproductive development (Feys et al., 1994; McConn and Browse, 1996). We suggest that these differences in the hormonal control of seed development in tomato and Arabidopsis may be related to differences in fruit type: siliques of Arabidopsis and many other Brassicaceae species are dehiscent dry fruit, whereas tomato produces a nondehiscent fleshy fruit.
The behavior of jai1 as a recessive sporophytic mutation (i.e., typical 3:1 Mendelian segregation) indicates that one or more COI1-dependent processes in maternal sporophytic tissue is required for seed maturation. It is possible that LeCOI1 regulates the synthesis of a vitamin or cofactor that requires transport from maternal tissues to the developing embryo, similar to the role of biotin synthesis in Arabidopsis embryogenesis (Patton et al., 1998). Alternatively, COI1 may promote the accumulation of proteins that supply reproductive tissues with sources of carbon or nitrogen. This hypothesis is consistent with the observation that the sterility of jai1 flowers is associated with a loss of accumulation of PI-II. This finding, together with the fact that tomato flowers contain high levels of endogenous JAs (Hause et al., 2000), demonstrates that PI expression in tomato reproductive tissues is regulated primarily by the JA/COI1 signaling pathway. It is likely that PI-II represents one of many proteins that are deficient in jai1 flowers. In this context, jai1 plants will provide a useful tool for identifying other COI1-dependent processes that operate in reproductive tissues and may be used as a genetic background in which to assess the functions of various JA/COI1-regulated genes in reproductive development.
In addition to a defect in the maternal control of seed development, jai1 plants also exhibited defects in male reproductive function. Most notable among these was a reduction in pollen viability and germination. It also was apparent that the stigma of jai1 flowers protruded from the anther cone of mature flowers, which is known to reduce pollination efficiency in some genotypes and under some environmental conditions (Rick and Dempsey, 1969). Although stigma exsertion did not appear to result from abnormal development of the anther cone, the tip of jai1 anther cones did exhibit localized browning and tissue collapse. Despite these anomalies in jai1 anther development, the mutant produced a sufficient amount of viable pollen to render it male fertile in genetic crosses (Li et al., 2001). Thus, we conclude that, unlike Arabidopsis, COI1 function is not absolutely required for male fertility in tomato.
Role of COI1 in Glandular Trichome Development and Anti-Herbivore Defense
Cultivated and wild species of tomato produce a variety of multicellular glandular and nonglandular trichomes that provide both physical and chemical (i.e., entrapment or toxicity) barriers against insect invaders (Duffey, 1986; Kennedy, 2002). In contrast to detailed knowledge about the molecular processes underlying the development of single-celled trichomes in Arabidopsis, relatively little is known about the genetic control of multicellular trichomes (Glover and Martin, 2000). Here, we provide genetic evidence that LeCOI1 performs an important role in the production of glandular trichomes on immature fruit and also modulates type-VI trichome density on leaves and sepals. Several questions concerning the effect of LeCOI1 on trichome development remain to be answered. For example, it is currently unclear whether the trichome phenotype of jai1 fruit (i.e., hairlessness) results from a defect in the same COI1-mediated process that affects trichome density on leaves and sepals. Because jai1 disrupts the developmental program leading to normal seed and fruit production, it is possible that the absence of fruit trichomes is a pleiotropic effect of abnormal fruit development. The normal growth and morphology of jai1 leaves and sepals, on the other hand, suggests that LeCOI1 plays a more specific role in modulating the development of type-VI trichomes in these tissues.
A role for LeCOI1 in promoting glandular trichome-based defense is supported by several observations. First, the loss of LeCOI1 function appears to affect glandular trichomes (e.g., type VI) but not the nonglandular trichome types. This finding is consistent with evidence indicating that different types of multicellular trichomes are under the control of different developmental programs (Glover and Martin, 2000). Second, because Arabidopsis trichomes are exclusively of the unicellular nonglandular type, a specific role for LeCOI1 in the development of glandular trichomes might explain why coi1 null mutants have not been reported to exhibit trichome-related phenotypes. Third, the reduced density of type-VI trichomes on jai1 leaves was correlated with a profound loss of resistance to two-spotted spider mites. This observation is in agreement with studies showing that type-VI trichomes mediate the effective resistance of wild tomato species to spider mites and that the defense of cultivated tomato to this herbivore is regulated by endogenous JAs derived from the octadecanoid pathway (Kennedy, 2002; Li et al., 2002a). Finally, a role for LeCOI1 in glandular trichome function is supported by the effect of jai1 on defense-related exudate chemistry, including the reduced accumulation of monoterpenes that exert defensive action against insect pests (Mahmoud and Croteau, 2002; Pichersky and Gershenzon, 2002). The monoterpene deficiency of jai1-1 fruit is explained readily by the complete lack of glandular trichomes on this tissue. The monoterpene content in jai1-1 sepals, however, appeared to be significantly less than that predicted simply from the reduced density of type-VI trichomes on mutant sepals. Thus, it is possible that LeCOI1 positively regulates both glandular trichome development and the production of monoterpenes that are stored in these structures. This interpretation is in agreement with recent studies showing that terpene emission from tomato leaves is stimulated by exogenous MeJA and herbivore attacks that activate JA signaling (Farag and Paré, 2002; Thaler et al., 2002).
It also is noteworthy that jai1 leaves are deficient in the MeJA-induced expression of PPO (Figure 1, Table 1), which constitutes >50% of the total protein in type-VI trichomes of tomato (Yu et al., 1992). The COI1-dependent expression of PPO is in agreement with previous studies showing that JA regulates PPO accumulation in tomato leaves, including the expression of specific PPO isoforms in type-VI trichomes (Constabel et al., 1995; Thipyapong et al., 1997; Thipyapong and Steffens, 1997). One interpretation of these observations is that type-VI trichome development is coordinated with the JA-regulated synthesis of compounds that are produced and stored in these glands. Consistent with this idea, Duffey (1986) observed a strong positive correlation between PPO activity and the density of type-VI trichomes in various Lycopersicon species and suggested that these patterns reflect genetic differences. The idea that biotic stress can influence the production of defense-related morphological structures fits with previous observations of increased trichome density in response to herbivory (Myers and Bazely, 1991; Karban and Baldwin, 1997) and the JA-induced formation of resin ducts involved in the synthesis of terpenoids in some conifers (Nagy et al., 2000; Martin et al., 2002; Hudgins et al., 2003). An important test of this hypothesis will be to determine whether changes in endogenous JA levels are sufficient to alter the development of defense-related morphological structures. The identification of LeCOI1 as a determinant of glandular trichome function indicates that the density and biosynthetic capacity of these secreting protuberances can be manipulated genetically through the altered expression of appropriate regulatory genes. Such an approach may be useful for enhancing plant resistance to herbivores or for increasing the production of the many trichome-derived natural products that have significant value to humans (Duke et al., 2000; Wang et al., 2001).
METHODS
Plant Material, Growth Conditions, and Isolation of jai1-1
Tomato (Lycopersicon esculentum) cv Micro-Tom was used as the “wild type” for all experiments except the cDNA microarray analysis, in which cv Castlemart was used as the wild type. Plants were grown in Jiffy peat pots (Hummert International, Earth City, MO) and maintained in growth chambers as described previously (Howe et al., 2000). Fast-neutron irradiation of Micro-Tom seeds was performed at the International Atomic Energy Agency (Seibersdorf, Austria). Calibrated doses between 12.7 and 17.8 Gy were used for mutagenesis, with the average dose being 15.9 Gy. M2 seeds were collected separately from 981 M1 plants. The appearance of visible phenotypes (chlorosis, variegation, albinism, and dwarfism) in one or more M2 plants from 161 (16%) M1 families indicated that the mutagenesis was effective.
Twenty-five M2 plants per M1 family were tested for a deficiency in methyl jasmonate (MeJA)–inducible polyphenol oxidase (PPO) and proteinase inhibitor II (PI-II) accumulation as follows. Approximately 120 18-day-old seedlings were enclosed in a Lucite box (10 × 32 × 60 cm) containing 5 μL of MeJA applied to cotton wicks that were spaced evenly within the box. Plants were exposed to MeJA vapor for 24 h and then incubated for an additional 24 h at ambient humidity in the absence of MeJA. A small piece of leaf tissue sampled from the lower leaf of each plant then was assayed for PPO activity as described previously (Howe and Ryan, 1999). Plants showing reduced PPO activity were tested immediately for PI-II accumulation using a radial immunodiffusion assay (Ryan, 1967; Zhao et al., 2003). The PI-II detection limit of this assay was ∼10 μg/mL tissue extract. From a total of 24,077 M2 plants tested, 18 putative mutants were identified that accumulated reduced levels of both PPO and PI-II in response to MeJA. Only one mutant (jai1-1; line 406A) exhibited undetectable PPO and PI-II accumulation. This line was backcrossed successively four times to both Micro-Tom and Castlemart. Micro-Tom and Castlemart were used as the recurrent pistillate parents in this series of crosses.
Selection of jai1-1 Homozygotes
Homozygous jai1-1 seedlings were selected from F2 populations as follows. Seeds were germinated on a piece of water-saturated filter paper in a closed Tupperware box in the dark at ambient temperature. After 4 to 5 days, when the emerging radical was ∼1 cm in length, the filter paper was resaturated with a solution of 1 mM MeJA. This solution was prepared by mixing 2 μL of pure MeJA (Bedoukian Research, Danbury, CT) with 75 μL of ethanol, followed by dilution into 10 mL of sterile distilled water. Seedlings were grown in the dark for an additional 24 to 36 h, at which time they were scored for sensitivity to MeJA on the basis of the phenotypes depicted in Figure 1C. jai1-1 homozygotes were transferred to peat pots and grown as described above. MeJA-sensitive seedlings (Jai1/Jai1 and Jai1/jai1-1) that were transferred to soil resumed normal growth and development.
A PCR-based assay was used to distinguish the jai1-1 deletion allele from the wild-type LeCoi1 allele. The assay used an upper primer (5′-GTGGAGACGATATGTTGAGACTAA-3′) that anneals to an intron-1 sequence present in both wild-type and jai1-1 genomic DNA. PCR amplification in combination with a second primer (5′-CCATGGAGTCCATCACCTAACAGT-3′) that anneals to a downstream intron-1 sequence (deleted in jai1-1 plants) gave a 525-bp PCR product corresponding to the wild-type allele. The PCR assay also used a third primer (5′-GTGGTCAGATCAGAGCCCTCTATT-3′) that anneals to a region downstream of the 3′ end point of the deletion and, in conjunction with the upstream primer, amplified a 777-bp fragment that is specific for jai1-1. PCR using genomic DNA from Jai1/jai1-1 heterozygotes resulted in the amplification of both the 525- and 777-bp products.
Spider Mite Bioassays
General procedures for rearing and handling two-spotted spider mites (Tetranychus urticae) were as described previously (Li et al., 2002a). Two-choice assays were conducted by placing 10 mites within a 1-cm circle located equidistant (1 cm) between single leaflets from 4- to 6-week-old wild-type and jai1-1 plants. A 10-cm Petri dish was placed over the arena to reduce ambient air currents and to prevent mites from escaping. All assays were performed at 26 ± 2°C at 40 to 60% RH and were terminated at 1 h after initiation of the trial. Fecundity assays involved the transfer of five adult female mites from bean leaves to wild-type or jai1-1 leaf discs (12 mm) that were placed on wetted cotton, as described by Rodriguez et al. (1971)(1972). Mites and eggs were counted at 24-h intervals.
Pollen Viability and Germination Assays
Freshly collected pollen was incubated in germination medium (10% sucrose, 100 mg/L boric acid, 300 mg/L calcium nitrate, 200 mg/L magnesium sulfate, and 100 mg/L potassium chloride) for 2 h at room temperature and then analyzed for pollen tube formation. Pollen tube length was recorded with a digital video camera (model MDS100; Kodak). Pollen grains were considered germinated if the tube length was greater than the diameter of the grain. The germination rate was calculated as the average germination percentage from 10 arbitrarily selected microscopic fields. Fluorescein diacetate/propidium iodide staining was used to measure pollen viability as described previously (Oparka and Read, 1994). Alexander's triple staining procedure (Alexander, 1980) was used to determine the proportion of pollen containing active cytoplasm.
Analysis of Trichomes
Scanning electron microscopy was performed with a JEOL 6400V scanning electron microscope (Tokyo, Japan) at an accelerating voltage of 15 kV. To examine the general pattern of trichome distribution on leaves, sepals, and green fruit, small pieces of tissue (5 × 5 mm) were fixed in 4% glutaraldehyde in 20 mM sodium phosphate buffer, pH 7.4, dehydrated through an ethanol series, critical point dried in CO2, and coated with gold using an EMSCOPE SC500 sputter coater (Ashford, UK). The density of type-VI trichomes on the adaxial surface of leaves and the lower surface of sepals (corresponding to the outer sepal surface for unopened flower buds) was determined by counting trichomes with a dissecting microscope equipped with a stage micrometer. All measurements were performed on wild-type and jai1-1 plants grown side by side in the same growth chamber. Because the trichome density of tomato leaflets is highly dependent on position along the developmental axis of the leaflet as well as leaf age, care was taken to survey comparable tissues of both genotypes.
Trichome exudates from fresh young leaves, sepals, and fruit were obtained by dipping the tissues in 3 mL of tert-butyl methyl ether (MTBE) for 5 min, with gentle shaking. MTBE-extracted compounds were concentrated under nitrogen gas and analyzed by gas chromatography–mass spectrometry as described previously (Lewinsohn et al., 2001). Monoterpene content was normalized to the surface area of the tissue used for each extraction. A stretched glass pipette was used to collect exudate directly from type-VI glands on wild-type fruit. Collected material was dissolved in MTBE, concentrated as described above, and analyzed by gas chromatography–mass spectrometry. The monoterpenes α- and β-pinene, limonene, and cis-β-ocimene were identified by comparison with authentic standards. Compounds corresponding to P5 and P6 (Figure 4) were not identified. Toluene was used as the internal standard for the quantification of monoterpenes.
Nucleic Acid Blot Analysis
RNA and DNA gel blot analyses were conducted as described by Li and Howe (2001). Duplicate RNA gels were stained with ethidium bromide to verify RNA quality and to ensure equal loading. A cDNA for tomato translation initiation factor eIF4A (cLED1D24) was used as the loading control. Three-week-old plants were enclosed in a Lucite box (60 × 32 × 17 cm) containing 2 μL of MeJA, which was dissolved in 0.25 mL of ethanol and distributed to several evenly spaced cotton wicks within the box. For each time point of sampling, five plants were removed from the box and leaf tissue was pooled for the extraction of RNA (Li and Howe, 2001).
cDNA Microarray Analysis
The microarray slide was composed of 607 tomato cDNAs that represent ∼500 unique tomato genes (Zhao et al., 2003). Each cDNA was spotted in triplicate on the slide. A complete list of the clones on the array is provided in the supplemental data online. Microarray analysis involved two experimental comparisons: (1) 18-day-old wild-type plants (cv Castlemart) treated with MeJA or mock-treated with ethanol; and (2) 18-day-old jai1-1 plants (in the Castlemart genetic background) treated with MeJA or mock-treated with ethanol. In each comparison, probes derived from MeJA-treated and ethanol-treated plants were considered as the test sample and reference sample, respectively. MeJA treatments were performed as described above. Mock-treated control plants were placed in a separate box in which an equivalent amount of ethanol was applied to cotton wicks. Plants were maintained in the box under standard growth conditions in the light for 8 h, after which time leaf tissue from six to eight plants of the same genotype was pooled for RNA isolation. Detailed procedures for probe preparation and labeling, hybridization and washing, and analysis of hybridization spot intensities are available at http://www.prl.msu.edu/howe.shtml.
Two biological replicate RNA samples were used for hybridization. Thus, four hybridization experiments were performed. In one of the two replicates, labeling of the two RNA samples with Cy5 or Cy3 dUTP was reversed to avoid potential dye-related differences in labeling efficiency. Spot intensities were quantified using GenePix Pro 3 image-analysis software (Axon, Foster City, CA). Ratio data were extracted and normalized to the set of spiking controls as described by Zhao et al. (2003). For data analysis, the ratios of the three replicate spots for each clone on the array were averaged. The ratio data for all 607 cDNAs on the array are provided in the supplemental data online. The final expression ratio was calculated as the average ratio from the two biological replicates (see supplemental data online). Only those genes with an expression ratio of >2.5 or <0.4 in both biological replicates were counted as being regulated differentially (see supplemental data online). The raw data for all hybridization experiments are available at http://www.prl.msu.edu/howe. shtml.
Cloning of LeCoi1
A BLAST (Basic Local Alignment Search Tool) search (Altschul et al., 1990) of the tomato EST database at The Institute for Genomic Research (http://www.tigr.org/) identified a single EST contig that was highly similar to Arabidopsis COI1 (TBLASTN score = 1085; E value = 2.1e-110). DNA sequence analysis of one clone (cLEB3P18) in this contig revealed a 1.1-kb insert that corresponds to the 3′ end of the cDNA. A combination of 5′ rapid amplification of cDNA ends (Gibco BRL) and Genome Walker (Clontech, Palo Alto, CA) technology was used to obtain additional sequence at the 5′ end of the cDNA, including ∼250 bp of DNA upstream of the putative ATG initiation codon. This sequence information was used to design gene-specific primers upstream (C1, 5′-CGGGATCCCTCTCCTCCATCTTCTTCAA-3′) and downstream (C2, 5′-CGAGCTCATACATATGGACAAGACACCT-3′); these were used to obtain a full-length cDNA by reverse transcription–PCR (RT-PCR). Reactions were performed with 5 μg of total RNA from Micro-Tom leaves and the Enhanced Avian HS RT-PCR-20 Kit (Sigma) as recommended by the manufacturer. The resulting 2.0-kb PCR product was ligated into pGEM-T vector (Promega) to generate plasmid pGEM-COI1. The cDNA insert (designated LeCoi1) was sequenced in its entirety and deposited in GenBank. The presence of an in-frame stop codon (TGA) 36 bp upstream of the initiator ATG indicates that the cDNA encodes a full-length polypeptide.
The genomic sequence of LeCoi1 was obtained from a BAC clone (249O9) that was identified by screening a tomato BAC library (Budiman et al., 2000) with EST clone cLEB3P18. To obtain the genomic sequence for LeCoi1, we designed PCR primers (based on the cDNA sequence) that amplify several overlapping fragments that span the locus. This sequence was assembled into a 9.4-kb contiguous sequence that was deposited in GenBank. Thermal asymmetric interlaced PCR (TAIL-PCR) (Liu and Whittier, 1995) was used to clone a fragment of DNA that spans the jai1-1 deletion end points and extends to the end of the 9.4-kb genomic sequence (Figure 8A). For this reaction, genomic DNA prepared from jai1-1 plants was used as a template for TAIL-PCR involving three gene-specific primers (C5, 5′-GAGGCAATATGTGGATTTGATGGA-3′; C6, 5′-CCACACCGTGTTCTTTTGAAGTGGA-3′; C7, 5′-GGAGACGATATGTTGAGACTAAGT-3′) that anneal to intron 1 of LeCoi1 and a degenerate “AD2” primer [5′-NGTCGA(G/C)(A/T)GANA(A/T)GAA-3′]. A 1.3-kb PCR fragment obtained from this reaction was cloned into pGEM-T and sequenced. Comparison of this sequence with the 9.4-kb genomic sequence identified the deletion end points.
Agrobacterium tumefaciens–Mediated Transformation
Plasmid pGEM-COI1 was digested with BamHI and SstI to release the 2.03-kb cDNA, which was cloned subsequently into BamHI and SstI sites of the binary vector pBI121 (Clontech). This construct was introduced into Agrobacterium tumefaciens strain AGLO and used to transform jai1-1 cotyledon explants as described previously (Li et al., 2001, 2003). Cotyledon tissue was obtained from jai1-1 homozygotes that were grown on sterile filter paper and selected from an F2 population using the root growth inhibition assay described above. The presence of the transgene in independent primary transformants (T1) was confirmed using the primer set C3 (5′-CTGCAAGTTAGGGCTGAAGATCTT-3′) and C4 (5′-GGCCAAGCACTTCCAATCCTCTAT-3′) in a PCR containing genomic DNA from each transformant. These primers amplify 1116- and 433-bp products corresponding to the endogenous LeCoi1 and 35S-LeCoi1 transgenes, respectively. Regenerated plants were transferred to the greenhouse for collection of T2 seeds. A PCR assay (see above) that detects both the jai1-1 deletion allele and the wild-type allele was used to confirm that regenerated T1 plants were homozygous for jai1-1.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact Gregg A. Howe, howeg@msu.edu.
Accession Numbers
The GenBank accession number for the LeCoi1 genomic sequence is AY423549. Other accession numbers are as follows: Arabidopsis COI1, NP_565919; rice COI1, BAB84399; tomato COI1, AY42350.
Supplementary Material
Acknowledgments
We thank Hui Chen for preliminary analysis of terpene profiles, Ewa Danielewicz at the Michigan State University Center for Advanced Microscopy for expert assistance with scanning electron microscopy, and Jonathan Vogel for assistance with cloning LeCoi1. We also thank David Shaffer and Josh Picotte for assistance with the mutant screening. Tomato EST clones and the tomato BAC library used in this study were obtained from the Clemson University Genomics Institute. This research was supported by grants from the National Institutes of Health (R01GM57795), the Michigan Life Science Corridor (085P1000466), the U.S. Department of Energy (DE-FG02-91ER20021), and the Michigan Agricultural Experiment Station at Michigan State University.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017954.
Footnotes
Online version contains Web-only data.
References
- Alexander, M.P. (1980). A versatile stain for pollen, fungi, yeast and bacteria. Stain Technol. 55, 13–18. [DOI] [PubMed] [Google Scholar]
- Allen, K.D. (2002). Assaying gene content in Arabidopsis. Proc. Natl. Acad. Sci. USA 99, 9568–9572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tool. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Andersson, B.A., Holman, R.T., Lundgren, L., and Stenhagen, G. (1980). Capillary gas chromatograms of leaf volatiles: A possible aid to breeders for pest and disease resistance. J. Agric. Food Chem. 28, 985–989. [Google Scholar]
- Beaudoin, N., and Rothstein, S.J. (1997). Developmental regulation of two lipoxygenase promoters in transgenic tobacco and tomato. Plant Mol. Biol. 33, 835–846. [DOI] [PubMed] [Google Scholar]
- Budiman, M.A., Mao, L., Wood, T.C., and Wing, R.A. (2000). A deep-coverage tomato BAC library and prospects towards development of an STC framework for genome sequencing. Genome Res. 10, 129–136. [PMC free article] [PubMed] [Google Scholar]
- Buttery, R.G., Ling, L.C., and Light, D.M. (1987). Tomato leaf volatile aroma components. J. Agric. Food Chem. 35, 1039–1042. [Google Scholar]
- Chao, W.S., Gu, Y.-Q., Pautot, V., Bray, E.A., and Walling, L.L. (1999). Leucine aminopeptidase RNAs, proteins, and activities increase in response to water deficit, salinity, and the wound signals systemin, methyl jasmonate, and abscisic acid. Plant Physiol. 120, 979–992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Conconi, A., Smerdon, M.J., Howe, G.A., and Ryan, C.A. (1996). The octadecanoid signalling pathway in plants mediates a response to ultraviolet radiation. Nature 383, 826–829. [DOI] [PubMed] [Google Scholar]
- Constabel, C.P., Bergey, D.R., and Ryan, C.A. (1995). Systemin activates synthesis of wound-inducible tomato leaf polyphenol oxidase via the octadecanoid defense signaling pathway. Proc. Natl. Acad. Sci. USA 92, 407–411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Creelman, R.A., and Mullet, J.E. (1997). Biosynthesis and action of jasmonates in plants. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 355–381. [DOI] [PubMed] [Google Scholar]
- Creelman, R.A., and Rao, M.V. (2002). The oxylipin pathway in Arabidopsis. In The Arabidopsis Book, C.R. Somerville and E.M. Meyerowitz, eds (Rockville, MD: American Society of Plant Biologists), doi/10.1199/tab.0009, http://www.aspb.org/publications/arabidopsis/. [DOI] [PMC free article] [PubMed]
- Devoto, A., Nieto-Rostro, M., Xie, D., Ellis, C., Harmston, R., Patrick, E., Davis, J., Sherratt, L., Coleman, M., and Turner, J.G. (2002). COI1 links jasmonate signaling and fertility to the SCF ubiquitin-ligase complex in Arabidopsis. Plant J. 32, 457–466. [DOI] [PubMed] [Google Scholar]
- Dombrowski, J.E. (2003). Salt stress activation of wound-related genes in tomato plants. Plant Physiol. 132, 2098–2107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duffey, S.S. (1986). Plant glandular trichomes: Their partial role in defence against insects. In Insects and the Plant Surface, S.R. Southwood and B. Juniper, eds (London: Edward Arnold), pp. 151–172.
- Duke, S.O., Canel, C., Rimando, A.M., Tellez, M.R., Duke, M.V., and Paul, R.N. (2000). Current and potential exploitation of plant glandular trichome productivity. Adv. Bot. Res. 31, 121–151. [Google Scholar]
- Ellis, C., and Turner, J.G. (2002). A conditionally fertile coi1 allele indicates cross-talk between plant hormone signalling pathways in Arabidopsis thaliana seeds and young seedlings. Planta 215, 549–556. [DOI] [PubMed] [Google Scholar]
- Farag, M.A., and Paré, P.W. (2002). C6-green leaf volatiles trigger local and systemic VOC emissions in tomato. Phytochemistry 61, 545–554. [DOI] [PubMed] [Google Scholar]
- Feys, B.J.F., Benedetti, C.E., Penfold, C.N., and Turner, J.G. (1994). Arabidopsis mutants selected for resistance to the phytotoxin coronatine are male-sterile, insensitive to methyl jasmonate, and resistant to a bacterial pathogen. Plant Cell 6, 751–759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gillaspy, G., Ben-David, H., and Gruissem, W. (1993). Fruits: A developmental perspective. Plant Cell 5, 1439–1451. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giovannoni, J. (2001). Molecular biology of fruit maturation and ripening. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 725–749. [DOI] [PubMed] [Google Scholar]
- Glover, B.J., and Martin, V. (2000). Specification of epidermal cell morphology. Adv. Bot. Res. 31, 193–213. [Google Scholar]
- Goossens, A., Häkkinen, S.T., Laakso, I., Seppänen-Laakso, T., Biondi, S., De Sutter, V., Lammertyn, F., Nuutila, A.M., Söderlund, H., Zabeau, M., Inzé, D., and Oksman-Caldentey, K.-M. (2003). A functional genomics approach toward the understanding of secondary metabolism in plant cells. Proc. Natl. Acad. Sci. USA 100, 8595–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hare, P.D., Seo, H.S., Yang, J.Y., and Chua, N.H. (2003). Modulation of sensitivity and selectivity in plant signaling by proteasomal destabilization. Curr. Opin. Plant Biol. 6, 453–462. [DOI] [PubMed] [Google Scholar]
- Hause, B., Stenzel, I., Miersch, O., Maucher, H., Kramell, R., Ziegler, J., and Wasternack, C. (2000). Tissue-specific oxylipin signature of tomato flowers: Allene oxide cyclase is highly expressed in distinct flower organs and vascular bundles. Plant J. 24, 113–126. [DOI] [PubMed] [Google Scholar]
- Hays, D.B., Wilen, R.W., Sheng, C., Moloney, M.M., and Pharis, R.P. (1999). Embryo-specific gene expression in microspore-derived embryos of Brassica napus: An interaction between abscisic acid and jasmonic acid. Plant Physiol. 119, 1065–1072. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He, Y., Fukushige, H., Hildebrand, D.F., and Gan, S. (2002). Evidence supporting a role of jasmonic acid in Arabidopsis leaf senescence. Plant Physiol. 128, 876–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heil, M., Koch, T., Hilpert, A., Fiala, B., Boland, W., and Linsenmair, K.E. (2001). Extrafloral nectar production of the ant-associated plant, Macaranga tanarius, is an induced, indirect, defensive response elicited by jasmonic acid. Proc. Natl. Acad. Sci. USA 98, 1083–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heitz, T., Bergey, D.R., and Ryan, C.A. (1997). A gene encoding a chloroplast-targeted lipoxygenase in tomato leaves is transiently induced by wounding, systemin, and methyl jasmonate. Plant Physiol. 114, 1085–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A., Lee, G.I., Itoh, A., Li, L., and DeRocher, A. (2000). Cytochrome P450-dependent metabolism of oxylipins in tomato: Cloning and expression of allene oxide synthase and fatty acid hydroperoxide lyase. Plant Physiol. 123, 711–724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howe, G.A., and Ryan, C.A. (1999). Suppressors of systemin signaling identify genes in the tomato wound response pathway. Genetics 153, 1411–1421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudgins, J.W., Christiansen, E., and Franceschi, V.R. (2003). Methyl jasmonate induces changes mimicking anatomical defenses in diverse members of the Pinaceae. Tree Physiol. 23, 361–371. [DOI] [PubMed] [Google Scholar]
- Ishiguro, S., Kawai-Oda, A., Ueda, K., Nishida, I., and Okada, K. (2001). The DEFECTIVE IN ANTHER DEHISCENCE1 gene encodes a novel phospholipase A1 catalyzing the initial step of jasmonic acid biosynthesis, which synchronizes pollen maturation, anther dehiscence, and flower opening in Arabidopsis. Plant Cell 13, 2191–2209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jacinto, T., McGurl, B., Franceschi, V., Delano-Freier, J., and Ryan, C.A. (1997). Tomato prosystemin promoter confers wound-inducible, vascular bundle-specific expression of the β-glucuronidase gene in transgenic tomato plants. Planta 203, 406–412. [Google Scholar]
- Karban, R., and Baldwin, I.T. (1997). Induced Response to Herbivory. (Chicago: University of Chicago Press).
- Kennedy, G.G. (2002). Tomato, pests, parasitoids, and predators: Tritrophic interactions involving the genus Lycopersicon. Annu. Rev. Entomol. 48, 51–72. [DOI] [PubMed] [Google Scholar]
- Kessler, A., and Baldwin, I.T. (2002). Plant responses to insect herbivory: The emerging molecular analysis. Annu. Rev. Plant Biol. 53, 299–328. [DOI] [PubMed] [Google Scholar]
- Kloek, A.P., Verbsky, M.L., Sharma, S.B., Schoelz, J.E., Vogel, J., Klessig, D.F., and Kunkel, B.N. (2001). Resistance to Pseudomonas syringae conferred by an Arabidopsis thaliana coronatine-insensitive (coi1) mutation occurs through two distinct mechanisms. Plant J. 26, 509–522. [DOI] [PubMed] [Google Scholar]
- Lee, G.I., and Howe, G.A. (2003). The tomato mutant spr1 is defective in systemin perception and the production of a systemic wound signal for defense gene expression. Plant J. 33, 567–576. [DOI] [PubMed] [Google Scholar]
- Lewinsohn, E., et al. (2001). Enhanced levels of the aroma and flavor compound S-linalool by metabolic engineering of the terpenoid pathway in tomato fruits. Plant Physiol. 127, 1256–1265. [PMC free article] [PubMed] [Google Scholar]
- Li, C., Liu, G., Xu, C., Lee, G.I., Bauer, P., Ling, H.-Q., Ganal, M.W., and Howe, G.A. (2003). The tomato Suppressor of prosystemin-mediated responses2 gene encodes a fatty acid desaturase required for the biosynthesis of jasmonic acid and the production of a systemic wound signal for defense gene expression. Plant Cell 15, 1646–1661. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, C., Williams, M.M., Loh, Y.-T., Lee, I.G., and Howe, G.A. (2002. a). Resistance of cultivated tomato to cell content-feeding herbivores is regulated by the octadecanoid-signaling pathway. Plant Physiol. 130, 494–503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, L., and Howe, G.A. (2001). Alternative splicing of prosystemin pre-mRNA produces two isoforms that are active as signals in the wound response pathway. Plant Mol. Biol. 46, 409–419. [DOI] [PubMed] [Google Scholar]
- Li, L., Li, C., and Howe, G.A. (2001). Genetic analysis of wound signaling in tomato: Evidence for a dual role of jasmonic acid in defense and female fertility. Plant Physiol. 127, 1414–1417. [PMC free article] [PubMed] [Google Scholar]
- Li, L., Li, C., Lee, G.I., and Howe, G.A. (2002. b). Distinct roles for jasmonic acid synthesis and action in the systemic wound response of tomato. Proc. Natl. Acad. Sci. USA 99, 6416–6421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li, X., Schuler, M.A., and Berenbaum, M.R. (2002. c). Jasmonate and salicylate induce expression of herbivore cytochrome P450 genes. Nature 419, 712–715. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., and Whittier, R.F. (1995). Thermal asymmetric interlaced PCR: Automatable amplification and sequencing of insert end fragments from P1 and YAC clones for chromosome walking. Genomics 25, 674–681. [DOI] [PubMed] [Google Scholar]
- Luckwill, L.C. (1943). The Genus Lycopersicon: Historical, Biological, and Taxonomic Survey of the Wild and Cultivated Tomatoes. (Aberdeen, Scotland: Aberdeen University Press).
- Mahmoud, S.S., and Croteau, R.B. (2002). Strategies for transgenic manipulation of monoterpene biosynthesis in plants. Trends Plant Sci. 7, 366–373. [DOI] [PubMed] [Google Scholar]
- Mandaokar, A., Kumar, V.D., Amway, M., and Browse, J. (2003). Microarray and differential display identify genes involved in jasmonate-dependent anther development. Plant Mol. Biol. 52, 775–786. [DOI] [PubMed] [Google Scholar]
- Martin, D.M., Gershenzon, J., and Bohlmann, J. (2002). Induction of volatile terpene biosynthesis and diurnal emission by methyl jasmonate in foliage of Norway spruce. Plant Physiol. 132, 1586–1599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McConn, M., and Browse, J. (1996). The critical requirement for linolenic acid is pollen development, not photosynthesis, in an Arabidopsis mutant. Plant Cell 8, 403–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Meissner, R., Jacobson, Y., Melamed, S., Levyatuv, S., Shalev, G., Ashri, A., Elkind, Y., and Levy, A. (1997). A new model system for tomato genetics. Plant J. 12, 1465–1472. [Google Scholar]
- Memelink, J., Verpoorte, R., and Kijne, J.W. (2001). ORCAnization of jasmonate-responsive gene expression in alkaloid metabolism. Trends Plant Sci. 6, 212–219. [DOI] [PubMed] [Google Scholar]
- Miersch, O., and Wasternack, C. (2000). Octadecanoid and jasmonate signaling in tomato (Lycopersicon esculentum Mill.) leaves: Endogenous jasmonates do not induce jasmonate biosynthesis. Biol. Chem. 381, 715–722. [DOI] [PubMed] [Google Scholar]
- Moura, D.S., Bergey, D.R., and Ryan, C.A. (2001). Characterization and localization of a wound-inducible type I serine-carboxypeptidase from leaves of tomato plants (Lycopersicon esculentum Mill.). Planta 212, 222–230. [DOI] [PubMed] [Google Scholar]
- Myers, J.H., and Bazely, D. (1991). Thorns, spines, prickles, and hairs: Are they stimulated by herbivory and do they deter herbivores? In Phytochemical Induction by Herbivores, D.W. Tallamy and M.J. Raupp, eds (New York: John Wiley), pp. 325–344.
- Nagy, N.E., Franceschi, V.R., Solheim, H., Krekling, T., and Christiansen, E. (2000). Wound-induced traumatic resin duct development in stems of Norway spruce (Pinaceae): Anatomy and cytochemical traits. Am. J. Bot. 87, 302–313. [PubMed] [Google Scholar]
- Oparka, K.J., and Read, N.D. (1994). The use of fluorescent probes for studies in living plant cells. In Plant Cell Biology: A Practical Approach, N. Harries and K.J. Oparka, eds (Oxford, UK: IRL Press), pp. 27–68.
- Park, J.H., Halitschke, R., Kim, H.B., Baldwin, I.T., Feldmann, K.A., and Feyereisen, R. (2002). A knock-out mutation in allene oxide synthase results in male sterility and defective wound signal transduction in Arabidopsis due to a block in jasmonic acid biosynthesis. Plant J. 31, 1–12. [DOI] [PubMed] [Google Scholar]
- Patton, D.A., Schetter, A.L., Franzmann, L.H., Nelson, K., Ward, E.R., and Meinke, D.W. (1998). An embryo-defective mutant of Arabidopsis disrupted in the final step of biotin synthesis. Plant Physiol. 116, 935–946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peña-Cortés, H., Willmitzer, L., and Sánchez-Serrano, J.J. (1991). Abscisic acid mediates wound induction but not developmental-specific expression of the proteinase inhibitor II gene family. Plant Cell 3, 963–972. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pichersky, E., and Gershenzon, J. (2002). The formation and function of plant volatiles: Perfumes for pollinator attraction and defense. Curr. Opin. Plant Biol. 5, 237–243. [DOI] [PubMed] [Google Scholar]
- Rao, M.V., Lee, H., Creelman, R.A., Mullet, J.E., and Davis, K.R. (2000). Jasmonic acid signaling modulates ozone-induced hypersensitive cell death. Plant Cell 12, 1633–1646. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rick, C.M., and Dempsey, W.H. (1969). Position of the stigma in relation to fruit setting of the tomato. Bot. Gaz. 130, 180–186. [Google Scholar]
- Rodriguez, J.G., Dabrowski, Z.T., Stoltz, L.P., Chaplin, C.E., and Smith, W.O. (1971). Studies on resistance of strawberries to mites. 2. Preference and nonpreference responses of Tetranychus urticae and T. turestani to water-soluble extracts of foliage. J. Econ. Entomol. 64, 383–386. [Google Scholar]
- Rodriguez, J.G., Knavel, D.E., and Aina, O.J. (1972). Studies in the resistance of tomatoes to mites. J. Econ. Entomol. 65, 50–53. [Google Scholar]
- Rotem, R., Fingrut, O., Moskovitz, J., and Flescher, E. (2003). The anticancer agent methyl jasmonate induces activation of stress-regulated c-Jun N-terminal kinase and p38 protein kinase in human lymphoid cells. Leukemia, 17, 2230–2234. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A. (1967). Quantitative determination of soluble cellular proteins by radial diffusion in agar gels containing antibodies. Anal. Biochem. 19, 434–440. [DOI] [PubMed] [Google Scholar]
- Ryan, C.A. (1973). Proteolytic enzymes and their inhibitors in plants. Annu. Rev. Plant Physiol. 24, 173–196. [Google Scholar]
- Ryan, C.A. (2000). The systemin signaling pathway: Differential activation of plant defensive genes. Biochim. Biophys. Acta 1477, 112–121. [DOI] [PubMed] [Google Scholar]
- Sanders, P.M., Lee, P.Y., Biesgen, C., Boone, J.D., Beals, T.P., Weiler, E.W., and Goldberg, R.B. (2000). The Arabidopsis DELAYED DEHISCENCE1 gene encodes an enzyme in the jasmonic acid synthesis pathway. Plant Cell 12, 1041–1062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaller, A., Bergey, D.R., and Ryan, C.A. (1995). Induction of wound response genes in tomato leaves by bestatin, an inhibitor of aminopeptidases. Plant Cell 7, 1893–1898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sivasankar, S., Sheldrick, B., and Rothstein, S.J. (2000). Expression of allene oxide synthase determines defense gene activation in tomato. Plant Physiol. 122, 1335–1342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Staswick, P.E. (1990). Novel regulation of vegetative storage protein genes. Plant Cell 2, 1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stenzel, I., Hause, B., Maucher, H., Pitzschke, A., Miersch, O., Ziegler, J., Ryan, C.A., and Wasternack, C. (2003). Allene oxide cyclase dependence of the wound response and vascular bundle-specific generation of jasmonates in tomato: Amplification in wound signalling. Plant J. 33, 577–589. [DOI] [PubMed] [Google Scholar]
- Stintzi, A., and Browse, J. (2000). The Arabidopsis male-sterile mutant, opr3, lacks the 12-oxophytodienoic acid reductase required for jasmonate synthesis. Proc. Natl. Acad. Sci. USA 97, 10625–10630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strassner, J., Schaller, F., Frick, U.B., Howe, G.A., Weiler, E.W., Amrhein, N., Macheroux, P., and Schaller, A. (2002). Characterization and cDNA-microarray expression analysis of 12-oxophytodienoate reductases reveals differential roles for octadecanoid biosynthesis in the local versus the systemic wound response. Plant J. 32, 585–601. [DOI] [PubMed] [Google Scholar]
- Thaler, J.S., Farag, M.A., Paré, P.W., and Dicke, M. (2002). Jasmonate-deficient plants have reduced direct and indirect defences against herbivores. Ecol. Lett. 5, 764–774. [Google Scholar]
- Thipyapong, P., Joel, D.M., and Steffens, J.C. (1997). Differential expression and turnover of the tomato polyphenol oxidase gene family during vegetative and reproductive development. Plant Physiol. 113, 707–718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thipyapong, P., and Steffens, J.C. (1997). Tomato polyphenol oxidase: Differential response of the polyphenol oxidase F promoter to injuries and wound signals. Plant Physiol. 115, 409–418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turner, J.G., Ellis, C., and Devoto, A. (2002). The jasmonate signal pathway. Plant Cell 14 (suppl.), S153.–S164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van der Hoeven, R., Ronning, C., Giovannoni, J., Martin, G., and Tanksley, S. (2002). Deductions about the number, organization, and evolution of genes in the tomato genome based on analysis of a large expressed sequence tag collection and selective genomic sequencing. Plant Cell 14, 1441–1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Villanueva, J., Canals, F., Prat, S., Ludevid, D., Querol, E., and Aviles, F.X. (1998). Characterization of the wound-induced metallocarboxypeptidase inhibitor from potato: cDNA sequence, induction of gene expression, subcellular immunolocalization and potential roles of the C-terminal propeptide. FEBS Lett. 440, 175–182. [DOI] [PubMed] [Google Scholar]
- Wang, E., Wang, R., DeParasis, J., Loughrin, J.H., Gan, S., and Wagner, G.J. (2001). Suppression of a P450 hydroxylase gene in plant trichome glands enhances natural-product-based aphid resistance. Nat. Biotechnol. 19, 371–374. [DOI] [PubMed] [Google Scholar]
- Wasternack, C., and Hause, B. (2002). Jasmonates and octadecanoids: Signals in plant stress responses and development. Prog. Nucleic Acid Res. Mol. Biol. 72, 165–221. [DOI] [PubMed] [Google Scholar]
- Weber, H. (2002). Fatty acid-derived signals in plants. Trends Plant Sci. 7, 217–224. [DOI] [PubMed] [Google Scholar]
- Weiler, E.W., Albrecht, T., Groth, B., Xia, Z.Q., Luxem, M., Lib, H., Andert, L., and Spengler, P. (1993). Evidence for the involvement of jasmonates and their octadecanoid precursors in the tendril coiling response of Bryonia dioica. Phytochemistry 32, 591–600. [Google Scholar]
- Wilen, R.W., van Rooijen, G.J.H., Pearce, D.W., Pharis, R.P., Holbrook, L.A., and Moloney, M.M. (1991). Effects of jasmonic acid on embryo-specific processes in Brassica and Linum oilseeds. Plant Physiol. 95, 399–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie, D.X., Feys, B.F., James, S., Nieto-Rostro, M., and Turner, J.G. (1998). COI1: An Arabidopsis gene required for jasmonate-regulated defense and fertility. Science 280, 1091–1094. [DOI] [PubMed] [Google Scholar]
- Xu, L.H., Liu, F.Q., Lechner, E., Genschik, P., Crosby, W.L., Ma, H., Peng, W., Huang, D.F., and Xie, D.X. (2002). The SCFCOI1 ubiquitin-ligase complexes are required for jasmonate response in Arabidopsis. Plant Cell 14, 1919–1935. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, H., Kowalski, S.P., and Steffens, J.C. (1992). Comparison of polyphenol oxidase expression in glandular trichomes of Solanum and Lycopersicon species. Plant Physiol. 10, 1885–1890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao, Y., Thilmony, R., Bender, C.B., Schaller, A., He, S.Y., and Howe, G.A. (2003). Virulence systems of Pseudomonas syringae pv. tomato promote bacterial speck disease in tomato by targeting the jasmonate signaling pathway. Plant J. 36, 485–499. [DOI] [PubMed] [Google Scholar]
- Ziegler, J., Keinänen, M., and Baldwin, I.T. (2001). Herbivore-induced allene oxide synthase transcripts and jasmonic acid in Nicotiana attenuata. Phytochemistry 58, 729–738. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.