Abstract
To test the hypothesis that caspase-like proteases exist and are critically involved in the implementation of programmed cell death (PCD) in plants, a search was undertaken for plant caspases activated during the N gene–mediated hypersensitive response (HR; a form of pathogen-induced PCD in plants) in tobacco plants infected with Tobacco mosaic virus (TMV). For detection, characterization, and partial purification of a tobacco caspase, the Agrobacterium tumefaciens VirD2 protein, shown here to be cleaved specifically at two sites (TATD and GEQD) by human caspase-3, was used as a target. In tobacco leaves, specific proteolytic processing of the ectopically produced VirD2 derivatives at these sites was found to occur early in the course of the HR triggered by TMV. A proteolytic activity capable of specifically cleaving the model substrate at TATD was partially purified from these leaves. A tetrapeptide aldehyde designed and synthesized on the basis of the elucidated plant caspase cleavage site prevented fragmentation of the substrate protein by plant and human caspases in vitro and counteracted TMV-triggered HR in vivo. Therefore, our data provide a characterization of caspase-specific protein fragmentation in apoptotic plant cells, with implications for the importance of such activity in the implementation of plant PCD.
INTRODUCTION
Programmed cell death (PCD), or apoptosis, is a basic genetically controlled process that functions in the development of multicellular organisms and in their responses to biotic and abiotic stresses. PCD provides a means to eliminate redundant, damaged, or infected cells. In mammals, apoptosis is recognized as a ubiquitous phenomenon with distinct morphological hallmarks, including condensation of the nucleus and the cytoplasm, cleavage of DNA into short 180-bp fragments (DNA laddering), and surface marking of the dying cells that are ingested by phagocytes (reviewed by Kerr et al., 1972). At the molecular level, there are two canonical pathways to apoptotic cell death. One involves the interaction of a death receptor with its ligand, and the second depends on the participation of mitochondria. The release of a number of factors—most notably cytochrome c—from the mitochondrial intermembrane space is regulated by proapoptotic and antiapoptotic members of the Bcl-2 family. Crosstalk between the two pathways exists (reviewed by Green, 1998; Adrain and Martin, 2001; Kaufmann and Hengartner, 2001). The end result of either pathway is the activation of caspases, a family of Cys proteases conserved evolutionarily from nematodes to humans (Thornberry and Lazebnik, 1998; Wolf and Green, 1999). Caspases are activated from dormant precursors in the course of apoptosis and introduce breaks after specific Asp (D) residues (hence their name) in a limited set of key cellular protein substrates. Caspases are among the most specific proteases known, and usually no more than one or two breaks per substrate protein molecule are introduced. Caspase-mediated protein fragmentation eventually leads to cell dismantling. The most prevalent executioner caspase in animal cells is caspase-3, which ultimately is responsible for the majority of the effects orchestrating cell death. In accordance with the pivotal role of caspases in PCD, caspase-specific peptide inhibitors based on the sequences cleaved by caspases and caspase gene knockouts are known to counteract apoptosis in animals (Ekert et al., 1999; Zheng et al., 1999).
In plants, several tissues or whole organs undergo cell death as part of their normal development, such as during senescence, or in response to pathogens and environmental stresses (reviewed by Greenberg, 1996). Whether or not common mechanisms underlie the implementation of PCD in animals and plants is an important emerging issue. Although the mechanisms of plant PCD are far less clear, several morphological and biochemical similarities between PCD in animals and plants have been described in different experimental systems, including condensation and shrinkage of the nucleus and cytoplasm, DNA laddering, and cytochrome c release from mitochondria (Danon et al., 2000; Balk and Leaver, 2001; Lam et al., 2001; Hoeberichts and Woltering, 2003). However, homologs to Bcl family genes, such as Bcl-2 and Bax, have not been shown to exist in plants, in spite of the complete sequencing of the Arabidopsis genome. Nevertheless, some studies have demonstrated that transgenic expression of such genes in plants can affect PCD pathways (Lacomme and Santa Cruz, 1999; Mitsuhar et al., 1999), and recently, a plant homolog of the animal Bax-Inhibitor1 gene was shown to be upregulated in response to wounding and pathogen challenge (Sanchez et al., 2000).
The case for the existence of caspases in plants is controversial. Although plant Cys proteolytic enzymes are known to be associated with PCD (Minami and Fukuda, 1995; D'Silva et al., 1998; Schmid et al., 1999; Solomon et al., 1999), no direct homologs of animal caspase genes have been identified in plants. However, some specific peptide inhibitors of animal caspases have been shown to affect the development of PCD. One of the examples of PCD in plants is the hypersensitive response (HR) that results from incompatible plant–pathogen interactions, which serves to prevent the spread of pathogens from the infection site (Greenberg, 1997; Heath, 2000). Specific inhibitors of animal caspase-1 and -3 (Ac-YVAD-cmk and Ac-DEVD-CHO, respectively) could attenuate bacteria- and Tobacco mosaic virus (TMV)–induced HR in tobacco leaves (del Pozo and Lam, 1998). Using synthetic fluorogenic caspase substrates, Ac-YVAD-AMC cleavage activity, but not Ac-DEVD-AMC cleavage activity, was detected in extracts prepared from tobacco plants undergoing TMV-induced HR (del Pozo and Lam, 1998). The same set of caspase inhibitors reduced the death of tobacco cells induced by isopentyladenosine (Mlejnek and Prochazka, 2002) and was efficient in the inhibition of cell death during menadion-induced apoptosis in tobacco protoplasts (Sun et al., 1999).
In Arabidopsis suspension cultures, nitric oxide– and H2O2-induced cell death was inhibited by the caspase-1 inhibitor (Clarke et al., 2000). Accordingly, an increase in caspase-1–like activity was found in cells treated with nitric oxide using a fluorogenic substrate (Clarke et al., 2000), whereas activation of caspase-3– and caspase-6– like proteases was observed during heat shock–induced apoptosis in tobacco suspension cells (Chen et al., 2000; Tian et al., 2000). PCD induced in tobacco cell suspension cultures by the ethylene-inducing xylanase fungal elicitor or by the protein kinase inhibitor staurosporine was blocked by the broad-range caspase inhibitors zVAD-fmk and BocD-fmk (Elbaz et al., 2002). Furthermore, the baculovirus antiapoptotic proteins p35 and IAP (inhibitor of apoptosis), which are known to inhibit animal caspases, also were efficient in preventing plant cell death induced by bacterial, fungal, and viral infections. In maize tissues, cocultivation with Agrobacterium spp resulted in a process resembling apoptosis that could be prevented by p35 and IAP (Hansen, 2000). Transgenic tobacco plants expressing the baculovirus Op-IAP were reported to acquire resistance to several necrotrophic fungal pathogens and to necrogenic Tomato spotted wilt virus (Dickman et al., 2001), whereas those expressing p35 showed partial inhibition of HR cell death upon bacteria and virus challenge (del Pozo and Lam, 2003). Finally, a family of distantly related caspase-like proteins termed metacaspases has been identified in the Arabidopsis genome (Uren et al., 2000). However, whether these predicted proteins possess caspase-specific proteolytic activity and are involved in the implementation of plant PCD remain to be demonstrated.
These observations suggest the existence of plant proteases (referred to here and below as plant caspases) with properties and cleavage specificities similar to those of animal caspases but perhaps being structurally different from them. However, for direct detection and analysis of such plant enzymes, it was crucial to identify a genuine plant protein substrate(s) that was (were) unknown.
Here, we used an approach based on the exploitation of the Agrobacterium tumefaciens–encoded VirD2 protein, which was predicted recently to be a possible substrate for caspases in our computer-assisted search for novel nuclear proteins subject to caspase fragmentation (N.V. Chichkova and A.B. Vartapetian, unpublished data). A. tumefaciens is a soil-borne plant pathogen capable of transferring DNA to the genome of higher plants, resulting in tumor formation. To initiate the process, VirD2 cleaves the agrobacterial tumor-inducing (Ti) plasmid DNA at a strictly defined sequence and becomes covalently attached to the newly formed 5′ DNA terminus (Yanofsky et al., 1986; Ward and Barnes, 1988; Durrenberger et al., 1989). Subsequent events lead to the formation of a single-stranded DNA fragment (T-DNA) linked to VirD2 and associated with other proteins. The migration of this complex to the plant cell nucleus is guided largely by the VirD2 nuclear localization signal (NLS) (Howard et al., 1992; Shurvinton et al., 1992; Ziemienowicz et al., 2001) and results in the integration of T-DNA into the plant genome and transformation of the host (Tinland et al., 1995). Because VirD2 operates in plant cells undergoing infection, this protein may be subjected to plant caspase fragmentation to protect the host from transformation. Therefore, in this work, VirD2 was used as a substrate to detect, partially purify, and characterize a putative plant caspase from tobacco plants triggered to undergo the HR by TMV infection. We demonstrate here that VirD2 fused with jellyfish green fluorescent protein (GFP) is cleaved by the identified plant caspase in vivo (during HR) and in vitro. A tetrapeptide aldehyde designed and synthesized on the basis of the plant caspase cleavage site prevented the fragmentation of the VirD2 substrate protein by the caspase in vitro and counteracted TMV-induced HR in vivo.
RESULTS
In Vitro Fragmentation of VirD2 by Human Caspase-3
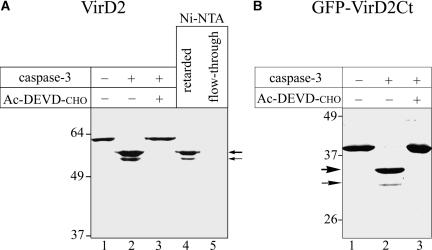
To test the ability of caspase-3, the principal human executioner caspase, to cleave the VirD2 protein, VirD2 containing an N-terminal six-His [(His)6] tag (VirD2-His) was expressed in Escherichia coli and purified further by nickel–nitrilotriacetic acid (Ni-NTA) agarose affinity chromatography. The purified preparation of the protein gave a single band in SDS-PAGE (Figure 1A, lane 1). Treatment of the VirD2-His protein with human caspase-3 resulted in the formation of two closely spaced bands with higher electrophoretic mobilities (Figure 1A, lane 2), indicating the terminal truncation of VirD2 at two different sites. Of note, the higher molecular mass VirD2 fragment was formed more efficiently (major component) than the lower molecular mass fragment (minor component). The caspase-3–specific inhibitor Ac-DEVD-CHO precluded the fragmentation of VirD2 (Figure 1A, lane 3). Both of the caspase-3 cleavage products could be retarded by the Ni-NTA agarose resin (Figure 1A, lane 4), indicating that they preserved the N-terminal (His)6 tag. Thus, both caspase-3 cleavage sites are located close to the C terminus of the VirD2 molecule.
Figure 1.
VirD2 Is a Target for Human caspase-3.
(A) Fragmentation of the full-length VirD2 protein. Recombinant full-length VirD2 (lane 1) was treated with caspase-3 in the absence (lane 2) or in the presence (lane 3) of Ac-DEVD-CHO inhibitor. The reaction mixtures were analyzed by 10% SDS-PAGE directly (lanes 1 to 3) or after fractionation by Ni-NTA agarose chromatography (lanes 4 and 5), followed by Coomassie blue staining.
(B) caspase-3 cleavage sites are located within the C-terminal portion of VirD2. GFP-VirD2Ct (lane 1) was treated with caspase-3 in the absence (lane 2) or in the presence (lane 3) of Ac-DEVD-CHO inhibitor. The reaction mixtures were analyzed by 12% SDS-PAGE.
Thick and thin arrows point to the major and minor caspase cleavage products, respectively. The positions of the molecular mass markers are indicated at left.
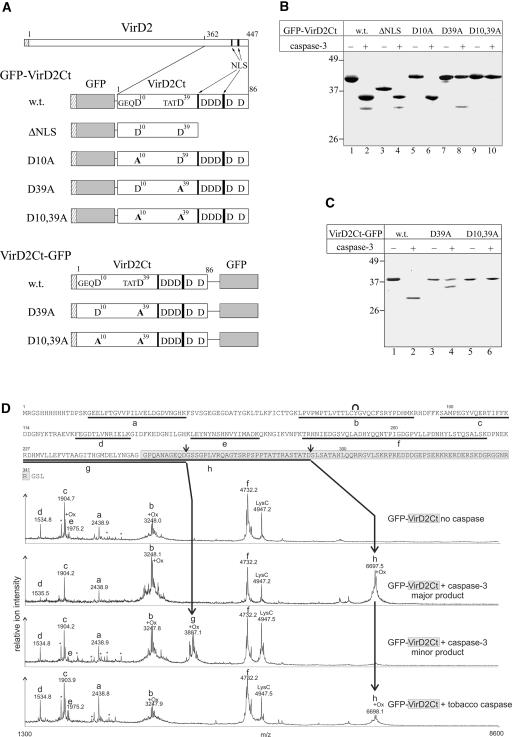
To construct a convenient protein substrate for further studies, the C-terminal 86–amino acid region of VirD2 (VirD2Ct), probably containing both of the caspase-3 cleavage sites, was fused to the C terminus of GFP and tagged with (His)6. The resulting GFP-VirD2Ct protein (Figure 2A, w.t.) then was confirmed to contain both of the sites recognized and cleaved by human caspase-3 in the entire VirD2 molecule (Figure 1B, lane 2).
Figure 2.
Identification of caspase-3 Cleavage Sites in VirD2.
(A) Scheme of VirD2 and VirD2Ct fused to GFP. Hatched rectangles at the N termini of recombinant proteins represent the (His)6 tags. The GFP moiety is shown in gray. Arrows point to two blocks of basic amino acids (black bars) representing the bipartite NLS of VirD2. All of the D residues present within VirD2Ct are indicated; those corresponding to the caspase cleavage sites are numbered and preceded by the upstream residues. Mutations are given in boldface. The relative sizes of the recombinant proteins are not shown to scale.
(B) Fragmentation of GFP-VirD2Ct wild-type (w.t.) and mutant proteins (designated as in [A]) with caspase-3.
(C) caspase-3–mediated fragmentation of VirD2Ct-GFP.
Reaction mixtures were resolved by 12% SDS-PAGE followed by Coomassie blue staining. The positions of the molecular mass markers are indicated at left.
(D) Mass spectrometric LysC peptide mapping of caspase cleavage sites. The complete amino acid sequence of the GFP-VirD2Ct protein is given at top. The VirD2Ct part of the sequence is highlighted in light gray. Peptide ions in the MALDI-TOF spectra are assigned to the GFP-VirD2Ct sequence as follows: residues 16 to 38 (peak a), 65 to 91 (peak b), 98 to 113 (peak c), 126 to 138 (peak d), 153 to 168 (peak e), 179 to 221 (peak f), 227 to 263 (peak g), and 227 to 292 (peak h). Background signals found in LysC digestions of control samples are marked with asterisks. The peak at m/z 4947.7 is the LysC autoproteolysis product that includes residues 156 to 203. The internal cyclization of the GFP chromophore in peptide b is symbolized with a curve above the sequence. Doublet signals that result from Met oxidation (+16 D) are marked with +Ox. Small arrows above the protein sequence indicate the identified caspase cleavage sites. The mass spectrum at bottom refers to the fragment generated by the plant caspase cleavage and is provided to permit its direct comparison with the spectra of caspase-3–generated fragments.
Within the C-terminal region of the VirD2 protein fused to the GFP are several D residues that could represent human caspase-3 cleavage sites (Figure 2A). To identify the two that are in fact involved in caspase-3 cleavage, a set of VirD2Ct mutants lacking one or more D residues were generated, fused to GFP, and tagged with (His)6 (Figure 2A). The recombinant proteins were produced in E. coli, purified, treated individually with caspase-3, and analyzed by SDS-PAGE (Figure 2B). The single D10A and D39A mutations resulted in the elimination of the minor, faster migrating product (Figure 2B, lane 6) and the major, slower migrating product (Figure 2B, lane 8), respectively, whereas the double mutant (D10,39A) had acquired complete resistance to caspase-3 hydrolysis (Figure 2B, lane 10). Elimination of all other downstream D residues had no effect on the cleavage pattern of GFP-VirD2Ct (Figure 2B, lane 4). Therefore, our results indicate that D39 and D10 represent the major and minor (suboptimal) caspase-3 cleavage sites, respectively, in VirD2.
To further support this conclusion, we inverted the order of the GFP and VirD2Ct moieties in the protein substrate to produce VirD2Ct-GFP (Figure 2A). Efficient caspase-3 cleavage of the VirD2Ct-GFP substrate at the proximal D39 site was observed (Figure 2C, lane 2), thus masking the cleavage at the distal D10 position. Substrate fragmentation at the suboptimal D10 site became evident only after mutating the D39 residue to A (Figure 2C, lane 4). As expected, the double mutant (D10,39A) was resistant to caspase-3 treatment (Figure 2C, lane 6). Thus, regardless of the order of the VirD2Ct and GFP moieties, human caspase-3 appears to cleave VirD2Ct preferentially at D39 and less efficiently at D10 (equivalent to D400 and D371, respectively, in the full-length VirD2).
To verify that the cleavage actually takes place at the expected residues, mass spectrometric (MS) characterization of caspase-3 cleavage sites in GFP-VirD2Ct was performed. Gel-purified GFP-VirD2Ct and the major and minor cleavage products generated by caspase-3 were subjected to enzymatic proteolysis using endoproteinase LysC. Digest peptides were captured on reversed-phase beads, desalted, and subjected to MS analysis on a linear-mode matrix-assisted laser-desorption ionization time-of-flight (MALDI-TOF) MS system. The C-terminal peptide at average mass-to-charge ratio (m/z) 6697.5 ± 0.8 was identified from the major (slower migrating) cleavage product (Figure 2D). The N terminus of this peptide h, expected at m/z 6697.3, is attributed to LysC cleavage within the GFP part of the substrate at K226, whereas its C terminus represents the caspase cleavage site at TATD292 (corresponding to TATD39 of the VirD2Ct part). The faster migrating band (Figure 1B, thin arrow) gave rise to the characteristic signal at m/z 3887.1 ± 0.8. The corresponding C-terminal peptide g (expected at m/z 3887.3) ends with the caspase-3 cleavage site at GEQD263 (corresponding to GEQD10 of the VirD2Ct part). None of these peptides was detected in LysC mapping experiments with intact GFP-VirD2Ct, although signals of several LysC-specific digest products (a to f) verified the identity of the protein (Figure 2D). Our analysis lacks the detection, from the intact GFP-VirD2Ct, of the expected LysC digest product at m/z 8531.4 (residues 227 to 309) that contains both caspase cleavage sites. Incomplete LysC cleavage in this C-terminal portion of the intact GFP-VirD2Ct or insufficient recovery of this big peptide might be responsible.
Therefore, we conclude that TATD39 and GEQD10 are the caspase-3 cleavage sites in GFP-VirD2Ct. It is noteworthy that both of the identified caspase-3 cleavage sites precede the bipartite NLS of VirD2, and as result, each cleavage of GFP-VirD2Ct produces a GFP-containing fragment devoid of the NLS. By contrast, the GFP-encompassing products of fragmentation of the alternative substrate, VirD2Ct-GFP, retain the NLS with either cleavage (for structures, see Figure 2A). This outcome was the key feature of our strategy to detect plant caspase activity in vivo.
Detection of a Plant Caspase-Like Activity in Tobacco Plants
Tobacco plants that possess the N gene are resistant to TMV and exhibit the HR after inoculation with that virus. In these plants, TMV is localized to the vicinity of the necrotic lesions formed at the late phases of the HR. The N gene–mediated response is temperature dependent, with the HR and resistance occurring only at temperatures <27°C (Kassanis, 1952). This feature of the N gene response allows plants to be inoculated and maintained at a temperature at which the resistance response is inoperative (33°C) before decreasing the temperature to initiate and synchronize the induction of HR (24°C). To assay a plant caspase activity induced by TMV-mediated HR, the GFP-VirD2Ct and VirD2Ct-GFP proteins were expressed as substrates in plants from a TMV-based vector, TMV(30B) (Shivprasad et al., 1999). For this purpose, nucleotide sequences encoding these proteins were inserted into TMV(30B) to give the hybrids TMV(GFP-VirD2Ct) and TMV(VirD2Ct-GFP), respectively.
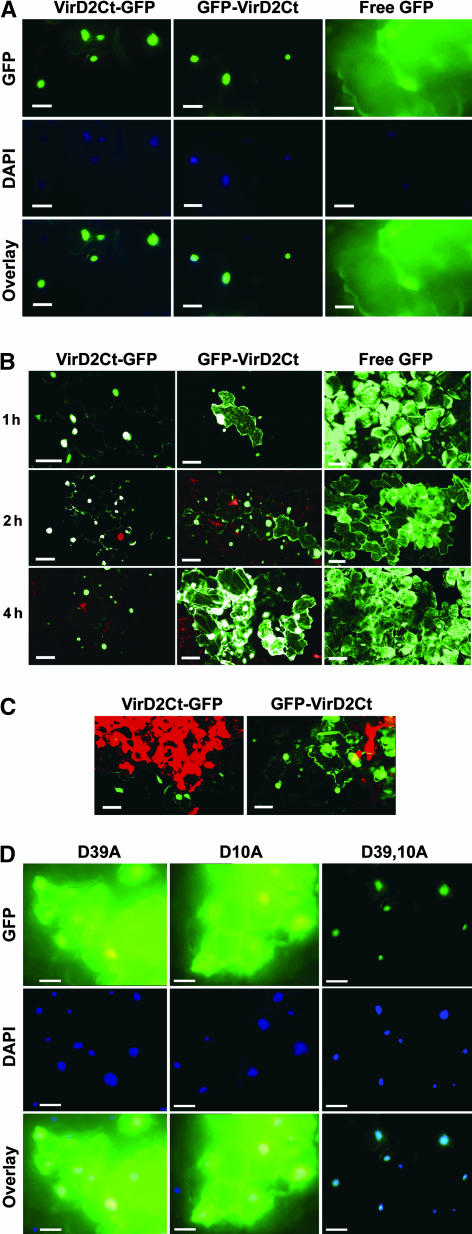
Two types of host plants were used for inoculation with the recombinant viruses: tobacco cv Samsun (with the nn genotype exhibiting systemic TMV infection [no HR] at both 24 and 33°C) and tobacco cv Samsun NN (with the NN genotype exhibiting temperature-dependent TMV-mediated HR as described above). The intracellular localization at the developing infectious sites of GFP-VirD2Ct and VirD2Ct-GFP was examined by confocal laser scanning microscopy and fluorescence microscopy. As expected, nuclear fluorescence was observed exclusively in the cells infected with TMV(GFP-VirD2Ct) or TMV(VirD2Ct-GFP) in the absence of the HR—that is, in Samsun NN plants at 33°C (Figure 3A) and in Samsun plants at either 33 or 24°C (data not shown). In a control experiment, infection with TMV expressing free GFP [TMV(GFP)] lacking the VirD2 sequence resulted in the distribution of GFP fluorescence throughout the cell (Figure 3A). This result indicated that no cleavage of the VirD2 NLS from GFP occurred in the absence of the HR. However, when the HR was induced by transferring Samsun NN plants from 33 to 24°C, rapid relocalization of fluorescence to the cytoplasm occurred in the case of TMV(GFP-VirD2Ct) (Figure 3B), presumably as a result of cleavage of the NLS from GFP.
Figure 3.
Intracellular Localization of VirD2Ct Derivatives Expressed as Fusions with GFP from a TMV(30B) Vector in Tobacco Samsun NN Plants during the HR.
(A) Fluorescence micrographs showing the nuclear localization of VirD2Ct-GFP and GFP-VirD2Ct in Samsun NN cells at 33°C (in the absence of HR). Free GFP expressed from a TMV(30B) vector was used as a control. 4′,6-Diamidino-2-phenylindole (DAPI) staining was used to visualize nuclei. The overlay images represent a superimposition of blue and green signals. Bars = 50 μm.
(B) Confocal images showing the intracellular localization of VirD2Ct-GFP and GFP-VirD2Ct in Samsun NN plants transferred from 33 to 24°C at 1, 2, and 4 h after transfer. Free GFP expressed from a TMV(30B) vector was used as a control. Bars = 50 μm.
(C) Confocal images showing the intracellular localization of VirD2Ct-GFP and GFP-VirD2Ct in Samsun NN at 24°C in a group of infected cells located at different distances from the center of a necrotic lesion (red). Bars = 50 μm.
(D) Fluorescence micrographs showing the intracellular localization of GFP-VirD2Ct containing single D10A and D39A and double D10,39A mutations in Samsun NN plants transferred from 33 to 24°C at 4 h after transfer. DAPI staining was used to visualize nuclei. The overlay images represent a superimposition of blue and green signals. Bars = 50 μm.
Cytoplasmic fluorescence became visible as early as 1 h after transfer, and at 4 h after transfer, it had spread throughout the entire patch of infected cells. By contrast, fluorescence in cells infected with TMV(VirD2Ct-GFP) was localized to the nucleus even in plants expressing the HR (Figure 3B). Interestingly, examination of a group of infected cells located at different distances from the center of the necrotic lesion revealed that, for TMV(GFP-VirD2Ct), the initial nuclear fluorescence of freshly infected cells farthest from the necrotic center was replaced by nuclear/cytoplasmic fluorescence in cells close to the necrotic area (Figure 3C), whereas for TMV(VirD2Ct-GFP), it remained confined to the nucleus (Figure 3C). These results suggested that the observed relocalization of the GFP-VirD2Ct fluorescent protein might be associated with its cleavage rather than with some general alteration in nuclear-cytoplasmic trafficking in the course of the HR. Considering the fact that human caspase-3 could cleave off the NLS sequences from the GFP moiety in GFP-VirD2Ct but not in VirD2Ct-GFP (see above), we suggest that a putative plant caspase with a similar specificity to human caspase-3 is activated during the HR in Samsun NN plants.
To further test this idea, the effects of the single D10A and D39A mutations and the double D10,39A mutation, corresponding to the cleavage sites of caspase-3 (Figures 2A and 2B), on the intracellular localization of GFP-VirD2Ct were assessed. These mutations were introduced individually into TMV(GFP-VirD2Ct) to give TMV(D10A), TMV(D39A), and TMV(D10,39A), respectively. Confocal laser scanning microscopy of infected sites on the Samsun NN plants inoculated with these viruses showed that all of the mutant GFP-VirD2Ct proteins accumulated efficiently in the nucleus at 33°C, as did the wild-type protein (data not shown). However, after the temperature was shifted to 24°C, both mutant proteins carrying single mutations (either D10A or D39A) became distributed throughout the cell (Figure 3D), whereas the protein carrying the double mutation (D10,39A) still retained its nuclear localization (Figure 3D, right column), indicating a lack of cleavage. This finding suggests that GFP-VirD2Ct is cleaved by a plant enzyme activated during TMV-mediated HR in vivo, at the same sites as those identified for caspase-3 in in vitro experiments (see above).
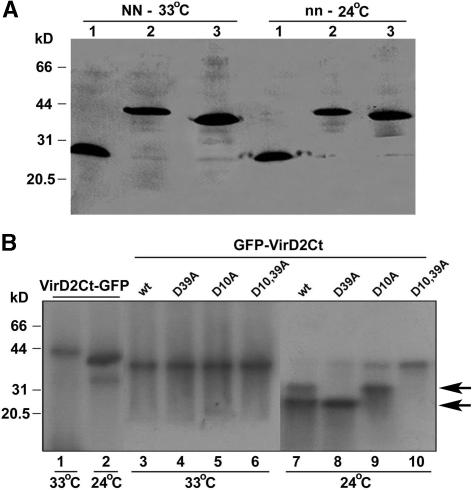
In parallel experiments, protein gel blot analysis using polyclonal antibody prepared against GFP showed that in the absence of the HR, either in Samsun plants at both 33 and 24°C (for 24°C, see Figure 4A) or in Samsun NN plants infected at 33°C (Figure 4A), the nonmutated recombinant proteins (VirD2Ct-GFP and GFP-VirD2Ct) were intact. A difference in size between VirD2Ct-GFP and GFP-VirD2Ct was caused by the presence of a short additional vector-derived sequence in VirD2Ct-GFP. However, when Samsun NN plants were shifted to 24°C to trigger the HR, fragmentation of these substrate fusion proteins at two distinct sites was evident (Figure 4B, lanes 2 and 7). Furthermore, under the same conditions, the single D10A or D39A mutation in GFP-VirD2Ct resulted in the elimination of the faster or the slower migrating product, respectively (Figure 4B, lanes 8 and 9). The relative size of the fragments formed was in agreement with the proposed pattern of proteolytic processing of VirD2. Most notably, the double mutation D10,39A prevented GFP-VirD2Ct from being cleaved even during the HR (at 24°C; Figure 4B, lane 10).
Figure 4.
Protein Gel Blot Analysis of the Recombinant VirD2Ct-GFP and GFP-VirD2Ct Proteins Expressed in Tobacco Plants from a TMV(30B) Vector at 33 or 24°C Using Antibody Prepared against GFP.
(A) Analysis of the nonmutated recombinant proteins (VirD2Ct-GFP [lanes 2] and GFP-VirD2Ct [lanes 3]) accumulated in Samsun NN plants at 33°C (NN-33°C) or in Samsun plants at 24°C (nn-24°C). A control sample isolated from TMV(GFP)-infected plants was loaded in lanes 1. The positions of the molecular mass markers are indicated at left.
(B) Analysis of nonmutated VirD2Ct-GFP (lanes 1 and 2), GFP-VirD2Ct (lanes 3 and 7), and its mutants D39A (lanes 4 and 8), D10A (lanes 5 and 9), and D10,39A (lanes 6 and 10) accumulated in Samsun NN plants either at 33°C or after transfer of plants to 24°C. The positions of the molecular mass markers are indicated at left. Arrows point to the caspase cleavage products. wt, wild type.
The results obtained are fully consistent with the notion of the activation, in the course of the HR, of a plant caspase(s) with the specificity of human caspase-3. In the wild-type protein substrate GFP-VirD2Ct, proteolytic cleavage at either D10 or D39 removes the NLS from the fluorescent protein, thus triggering its relocalization from the nucleus. Accordingly, the single D/A mutations do not rescue the nuclear accumulation of the cleavage product. However, when both of the cleavage sites are mutated simultaneously, the substrate protein acquires resistance to cleavage by a putative plant caspase and preserves the nuclear localization.
Partial Purification and Properties of a Plant Caspase
Leaves of N gene–carrying tobacco Xanthi nc plants (with the NN genotype) were inoculated with wild-type TMV, incubated at 33°C for 2 days, transferred to 24°C to induce the HR until many necrotic lesions became clearly visible, and homogenized. The extracts thus obtained were subjected to two DEAE cellulose chromatography steps, and fractions were assayed for the ability to cleave specifically the exogenously added GFP-VirD2Ct protein substrate (expressed in and purified from E. coli as described above). The reaction mixtures were fractionated by SDS-PAGE using the GFP-VirD2Ct substrate treated with human caspase-3 as a control. In fractions eluted from the column at 70 to 100 mM NaCl, proteolytic activity was detected, giving a protein fragment similar in electrophoretic mobility to that produced by caspase-3 cleavage at D39 (Figure 5, lanes 2 and 3). The peak fraction from the second chromatography step did not appear to contain contaminating protease activities that interfere with analysis; therefore, it was selected for the further characterization of the putative plant caspase.
Figure 5.
Tobacco Caspase-Like Protease Cleaves GFP-VirD2Ct at D39.
Wild type (w.t.; lanes 1 to 3) and D39A mutant (lane 4) GFP-VirD2Ct were treated with caspase-3 (lane 2) or with the partially purified plant enzyme (lanes 3 and 4). The samples were fractionated as described for Figure 2, and the proteins were detected by Coomassie blue staining. The arrow points to the plant caspase cleavage product.
The D39A mutation in GFP-VirD2Ct abrogated substrate fragmentation by the plant enzyme (Figure 5, lane 4). Furthermore, mass spectrometric analysis of the gel-purified and LysC-digested GFP-VirD2Ct cleavage product generated by plant caspase permitted the identification of the same C-terminal peptide at average m/z 6698.1 ± 0.8 (Figure 2D, bottom) as the peptide originating from the caspase-3 major cleavage product. Thus, plant caspase cleaves GFP-VirD2Ct at TATD39, indicating that the substrate specificity of the plant caspase is similar in part to that of the human caspase. However, the partially purified plant caspase reproducibly failed to hydrolyze the substrate at GEQD10, which is the suboptimal cleavage site for human caspase-3. Therefore, we suggest that yet another caspase-like enzyme may become activated in plant cells that is directly responsible for the D10 cleavage and the relocalization in vivo of GFP-VirD2Ct containing the mutation D39A.
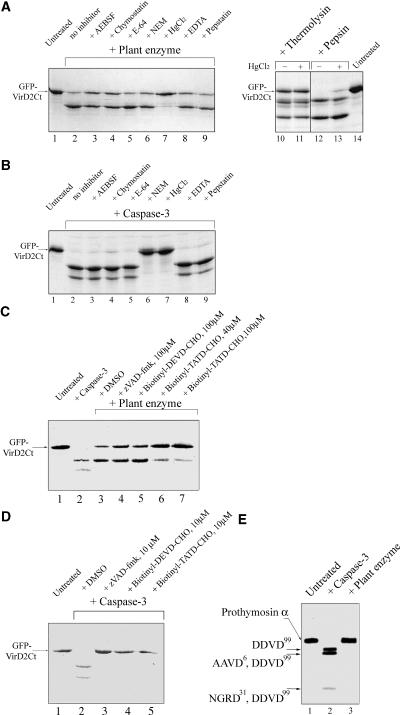
To determine whether the plant caspase-like enzyme under study is likely to be a Cys proteinase, as animal caspases are, inhibition of GFP-VirD2Ct cleavage with various protease inhibitors was attempted. Ser proteinase [4-(2-aminoethyl)benzenesulfonyl fluoride and chymostatin], aspartyl proteinase (pepstatin), and metallo proteinase (EDTA) inhibitors were ineffective at inactivating the plant enzyme (Figure 6A). Of the Cys protease inhibitors tested, mercuric chloride abolished the substrate fragmentation by both plant and human enzymes, N-ethylmaleimide inhibited human enzyme but not the plant enzyme, and neither of the enzymes was inhibited by E-64 (Figures 6A and 6B). Notably, mercuric chloride under the conditions used failed to inhibit thermolysin and pepsin, a metallo protease and an aspartyl protease, respectively (Figure 6A, lanes 10 to 13). These results indicate the specificity of the inhibitory action of mercuric chloride and support the notion that the plant caspase-like enzyme possesses a Cys residue essential for its catalytic activity.
Figure 6.
Comparison of the Specificity of Tobacco Caspase with That of Human caspase-3.
(A) and (B) Testing of Ser protease, Cys protease, aspartyl protease, and metallo protease inhibitors for abrogation of the GFP-VirD2Ct cleavage by plant caspase (A) and human caspase-3 (B). Lanes 10 to 14 in (A) indicate that mercuric chloride at 200 μM does not prevent the thermolysin- and pepsin-mediated degradation of the substrate protein. AEBSF, 4-(2-aminoethyl)benzenesulfonyl fluoride; NEM, N-ethylmaleimide.
(C) Biotinyl-TATD-CHO specifically inhibits the plant enzyme-mediated cleavage of GFP-VirD2Ct, whereas zVAD-fmk and biotinyl-DEVD-CHO do not. Concentrations of the inhibitors are indicated.
(D) Inhibition of the caspase-3–mediated fragmentation of GFP-VirD2Ct by 10 μM peptide aldehydes.
(E) Human prothymosin α containing multiple caspase-3 cleavage sites (lane 2) is resistant to fragmentation with tobacco caspase (lane 3). The caspase-3 cleavage sites leading to the generation of the prothymosin α fragments (Evstafieva et al., 2003) are indicated at left.
The samples in (A) to (D) were analyzed by 12% SDS-PAGE followed by Coomassie blue staining, and those in (E) were fractionated on an 8% polyacrylamide/7 M urea gel and visualized by methylene blue staining.
To compare in more detail the plant caspase activity with that of human caspase-3, a set of peptide-based inhibitors known to inactivate animal caspases was tested. Surprisingly, neither zVAD-fmk nor biotinyl-DEVD-CHO could inhibit the plant enzyme even at 100 μM (Figure 6C, lanes 4 and 5), whereas human caspase-3 hydrolysis was abolished completely by these inhibitors at 10 μM (Figure 6D, lanes 3 and 4). Based on the sequence of the plant caspase cleavage site in VirD2, TATD39 (identified above; see also Figure 2A), a potential inhibitor of plant caspase, biotinyl-TATD-CHO, was designed and synthesized. This biotinylated peptide aldehyde inhibited plant caspase at inhibitor concentrations of ≥40 μM (Figure 6C, lanes 6 and 7). It also proved to be a human caspase-3 inhibitor, with hydrolysis being prevented completely at 10 μM (Figure 6D, lane 5).
Therefore, we conclude that the plant caspase under study has greater substrate specificity than does human caspase-3. To further substantiate this conclusion, we tested an unrelated human protein, prothymosin α, for plant caspase fragmentation. Despite its small size, this protein possesses three rather different recognition sites for human caspase-3 cleaved with different efficiencies: DDVD99 > AAVD6 > NGRD31 (Evstafieva et al., 2003). However, the plant caspase could not cleave prothymosin α at any of these sites (Figure 6E), supporting the conclusion that the plant enzyme and human caspase-3 have partially overlapping but distinct substrate preferences.
Effect of Plant Caspase Inhibitor on Cell Death Mediated by TMV Infection
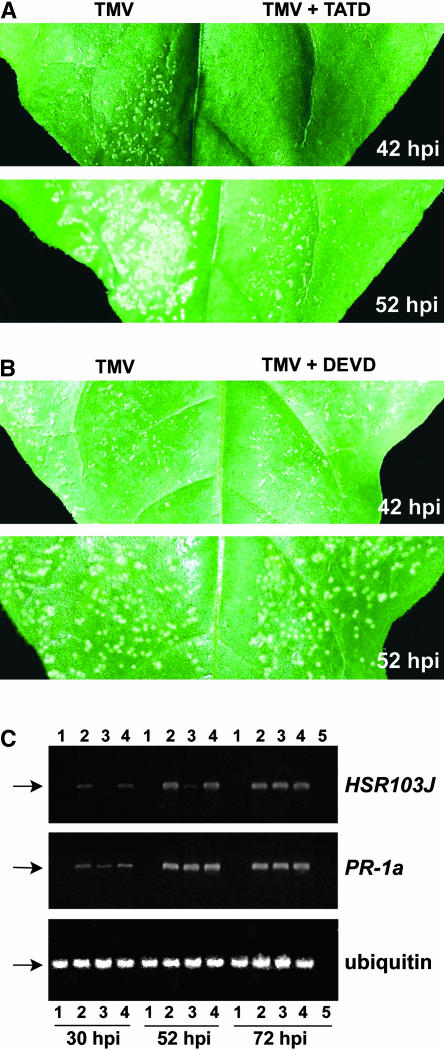
To determine whether the newly discovered plant caspase-like enzyme activated in the course of the TMV-mediated HR could be related to the occurrence of PCD, the effect of the specific inhibitor biotinyl-TATD-CHO on the necrotization of tobacco Samsun NN leaves upon TMV infection was tested. We coinoculated tobacco (Samsun NN) leaves with TMV(30B) and biotinyl-TATD-CHO at 24°C using diluent (instead of the peptide) as a control. Although characteristic symptoms of HR, such as the formation of necrotic lesions, first occurred at ∼30 h after inoculation and became well developed at 42 h after inoculation in the control (diluent-coinoculated) half-leaves (Figures 7A and 7B, 42 hpi), no necrotic lesions were detected in the biotinyl-TATD-CHO–inoculated half-leaves at this time (Figure 7A, 42 hpi). At 52 h after inoculation, necrotic lesions became visible on the biotinyl-TATD-CHO–inoculated half-leaves, their average size (diameter) being typically twofold to threefold smaller than those in the control half-leaves (Figure 7A, 52 hpi). At later time points, the difference between the lesion sizes disappeared. However, the rates of accumulation of TMV measured by ELISA were similar in biotinyl-TATD-CHO–inoculated and diluent-inoculated tissues at all time points up to 72 h after inoculation (Table 1), suggesting that biotinyl-TATD-CHO is involved in the inhibition of TMV-mediated HR rather than in the suppression of TMV accumulation.
Figure 7.
Effect of the Caspase-Specific Inhibitors (1 mM) Biotinyl-TATD-CHO and Biotinyl-DEVD-CHO on the HR Cell Death in Tobacco Samsun NN Plants Infected with 0.1 μg/μL TMV.
(A) and (B) Effect of biotinyl-TATD-CHO (TATD; [A]) or biotinyl-DEVD-CHO (DEVD; [B]) on the formation of necrotic lesions at 42 and 52 h after inoculation (hpi). The inhibitors were coinoculated mechanically with a TMV preparation. Control half-leaves were coinoculated with diluent.
(C) Reverse transcriptase–mediated (RT) PCR analysis showing the effect of biotinyl-TATD-CHO or biotinyl-DEVD-CHO on HSR203J and PR-1a transcription at 30, 52, and 72 h after inoculation. Ethidium bromide–stained agarose gels show typical RT-PCR products corresponding to the fragments of HSR203J mRNA (383 bp), PR-1a mRNA (300 bp), and ubiquitin mRNA (176 bp) used as a control, as indicated by the arrows. Lanes 1 to 4 represent samples from mock-inoculated plants (lanes 1) or TMV-inoculated plants in the absence of caspase inhibitors (lanes 2) or in the presence of biotinyl-TATD-CHO (lanes 3) or biotinyl-DEVD-CHO (lanes 4). Lanes 5 represent the control (sample infected with TMV in the absence of caspase inhibitors at 72 h after inoculation), in which the RT reaction mix, without reverse transcriptase, was used as a template. Presented here are PCR results from 35 cycles corresponding to the log-linear phase of amplified PCR products of both HSR203J and PR-1a in samples infected with TMV in the absence of caspase inhibitors.
Table 1.
Accumulation of TMV in Inoculated Leaves of N. tabacum Samsun NN Plants in the Presence or Absence of Caspase Inhibitors
| Amount of TMV Antigen (μg/g leaf)
|
||||
|---|---|---|---|---|
| Inhibitor | 30 HAIa | 42 HAI | 52 HAI | 72 HAI |
| None | 15 ± 3 | 59 ± 7 | 80 ± 6 | 101 ± 9 |
| Biotinyl-TATD-CHO | 17 ± 4 | 64 ± 5 | 78 ± 8 | 98 ± 7 |
| Biotinyl-DEVD-CHO | 13 ± 5 | 60 ± 6 | 84 ± 6 | 95 ± 8 |
Accumulation was determined by ELISA using serial dilutions of TMV preparation as concentration standards. Data shown are means ± se of three independent experiments with three replicates each.
HAI, hours after inoculation.
To confirm this suggestion at the molecular level, we examined the effect of biotinyl-TATD-CHO on the expression of a plant gene, HSR203J, an early marker of HR cell death, whose expression is activated several hours before the appearance of necrotic lesions induced by pathogens (Pontier et al., 1994). Semiquantitative reverse transcriptase–mediated PCR showed that in the presence of biotinyl-TATD-CHO, HSR203J RNA transcripts accumulated with a delay (they were not detected at 30 h after inoculation; Figure 7C, lanes 3 versus lanes 2) and to significantly lower levels, being reduced by >85% compared with samples inoculated with TMV in the absence of biotinyl-TATD-CHO (Figure 7C, lanes 3 versus lanes 2) at 52 h after inoculation. At later time points (72 h after inoculation), the difference in the expression of HSR203J disappeared (Figure 7C, lanes 3 versus lanes 2). The levels of ubiquitin mRNA used as a constitutively expressed internal control with or without biotinyl-TATD-CHO were similar in all samples (Figure 7C, ubiquitin). Thus, the inhibition of the necrotization of Samsun NN leaves upon TMV infection by biotinyl-TATD-CHO was correlated with a delay in the activation of HSR203J expression. By contrast, the TMV-mediated activation of the PR-1a gene (a common marker for the defense response) was suppressed only slightly by biotinyl-TATD-CHO at 30 h after inoculation and was not affected at all at later stages (Figure 7C, lanes 3 versus lanes 2).
All attempts to suppress the formation of necrotic lesions in TMV-infected tobacco Samsun NN leaves by coinoculation of biotinyl-DEVD-CHO, which did not inhibit the plant caspase in vitro (see above), also were unsuccessful (Figure 7B). Nor did biotinyl-DEVD-CHO affect the multiplication and spread of TMV in these plants or TMV-induced HSR203J or PR1-1a gene expression (Figure 7C, lanes 4 versus lanes 2). We conclude that the effect of biotinyl-TATD-CHO on the development of the TMV-mediated HR is highly specific. Although the plant caspase inhibitor biotinyl-TATD-CHO could not completely abolish the TMV-induced HR, it markedly retarded the appearance of the HR cell death. However, this partial inhibition of the HR did not result in the systemic spread of TMV (data not shown).
DISCUSSION
A large body of circumstantial evidence, based primarily on the use of mammalian caspase inhibitors, suggested that plant caspases might exist and be involved in the implementation of the PCD in various plant systems. However, these proteolytic enzymes remained elusive, mainly because natural protein targets of putative plant caspases were not known, and no reliable way to find them existed. Nor were their cleavage site requirements known, so research was based on analogy with animal caspase cleavage sites.
In our work, the VirD2 protein of A. tumefaciens was used for the detection, identification, and partial purification of a tobacco caspase, based on our prediction that this protein might represent a genuine caspase target. Indeed, we demonstrated that VirD2 could be cleaved by human caspase-3 at one major and one suboptimal site within the C-terminal portion of VirD2, upstream of its NLS. Moreover, because VirD2 is active in plant cells, this finding provided grounds for a search for a plant caspase-like activity capable of VirD2 processing at identical sites.
To classify the plant enzyme as an analog of animal caspases, we propose four major criteria: (1) this protein should be a protease; (2) its substrate specificity should be similar to that of an animal caspase; (3) it should become activated in the course of plant PCD; and (4) specific inhibitor(s) of this enzyme should counteract plant PCD. The plant enzyme under study meets all four of these criteria.
In the first approach, VirD2 was expressed as N- or C-terminal fusion proteins with GFP from a TMV vector. In these experiments, TMV also played the role of inducer of the HR. When the HR was induced, rapid relocalization of the fluorescence to the cytoplasm occurred in TMV(GFP-VirD2Ct), presumably because the NLS became detached from GFP-infected plants, whereas fluorescence in cells infected with TMV(VirD2Ct-GFP) remained localized to the nucleus. Given the fact that human caspase-3 also could cleave off the NLS sequences from the GFP moiety in GFP-VirD2Ct but not in VirD2Ct-GFP, it seemed possible that some putative plant caspase with a similar specificity is activated during the HR in tobacco plants. Mutational analysis of potential cleavage sites has identified two sites at which GFP-VirD2Ct is cleaved by a plant enzyme activated during TMV-mediated HR in vivo (TATD and GEQD) that are identical to those identified for caspase-3 in the experiments in vitro.
In the second approach, we used VirD2 as a substrate to test plant caspase activity induced during the HR in vitro. This approach allowed us to partially purify the enzyme and confirm its cleavage specificity.
In the third approach, the tetrapeptide aldehyde biotinyl-TATD-CHO, designed and synthesized on the basis of the elucidated plant caspase cleavage site, was shown to prevent the fragmentation of VirD2 by plant and human caspases in vitro and to counteract TMV-triggered HR in vivo.
Thus, our findings provide strong in vivo and in vitro evidence that a caspase-like protease is activated in tobacco plants during the N gene–mediated HR triggered by TMV infection. However, because no direct homologs to animal caspases have been found in plants in database searches, the plant caspase identified in this work presumably represents a functional analog of animal caspases.
The tobacco caspase described in this work appears to possess even greater specificity than human caspase-3 because (1) the plant enzyme hydrolyzes only a single peptide bond in the substrate protein, corresponding to the major cleavage site of human caspase; (2) of several caspase-targeted peptide inhibitors tested (zVAD-fmk, Ac-DEVD-CHO, biotinyl-DEVD-CHO, and biotinyl-TATD-CHO), all were efficient in caspase-3 inhibition, whereas the plant enzyme was inhibited only by biotinyl-TATD-CHO synthesized on the basis of the plant caspase cleavage site; and (3) plant caspase failed to process a model protein, prothymosin α, which possesses several very different sites for caspase-3.
The biotinyl-TATD-CHO concentration required to inhibit the plant enzyme was severalfold greater than that required to inhibit human caspase-3. A possible explanation for this difference is that a longer peptide moiety is optimal for recognition by plant caspase protease. Animal caspase-2, for example, requires pentapeptide substrates for efficient recognition (Talanian et al., 1997), although this appears to be an exception for animal caspases.
Our findings also suggest that in plants, as in animals, a family of caspases may exist, of which the enzyme identified is one of the members. Indeed, to prevent the proteolytic processing and nuclear escape of GFP-VirD2Ct substrate during the HR in tobacco leaves, mutation of both of the sites cleaved by human caspase-3, TATD39 and GEQD10, was required, whereas the plant enzyme under study was strictly specific to the former site. We infer from this observation that another plant caspase with a recognition sequence matching GEQD may become activated in the course of plant PCD. Consistent with this notion, the use of a set of peptide inhibitors targeting different members of the plant caspase family may lead to a more pronounced suppression of the HR in plants than did biotinyl-TATD-CHO alone.
Our observations show that the synthetic peptide inhibitor for caspase, biotinyl-TATD-CHO, dramatically retards the development of necrotic lesions, the macroscopic symptoms of cell death mediated by TMV. The fact that the suppression by biotinyl-TATD-CHO of the induction of TMV-triggered cell death was correlated with a delay in the activation of the HSR203J gene, an early HR-specific molecular marker, indicates that the plant caspase described here regulates an early step in the HR preceding the activation of HSR203J. By contrast, the finding that the expression of the PR-1a gene (a marker for the host defense response during HR) was affected only slightly by biotinyl-TATD-CHO is consistent with the idea that host defense mechanisms and HR cell death may be regulated by different pathways that are expressed coordinately (del Pozo and Lam, 1998; Lam and del Pozo, 2000).
Interestingly, del Pozo and Lam (1998) found that another animal caspase inhibitor, Ac-DEVD-CHO, suppressed the development of bacteria-induced HR symptoms (formation of necrotic lesions) in plants as well as the activation of its early gene markers (such as HSR203J and HIN1). Although under our assay conditions, in which TMV-triggered HR cell death was suppressed by biotinyl-TATD-CHO, Ac-DEVD-CHO had no effect (see above), we suggest that under different experimental (or physiological) conditions, different plant caspases may be activated. Of note, a transgenically expressed physiological inhibitor of animal cell death, the baculovirus p35 protein, suppressed plant PCD only partially and did not affect the formation of necrotic lesions (del Pozo and Lam, 2003), possibly as a result of the low levels of p35 produced in transgenic plants. At the same time, continual expression of this protein in transgenic plants allowed the virus to overcome the HR and spread systemically (del Pozo and Lam, 2003). By contrast, the transient inhibition of the HR by synthetic peptides (which are probably unstable in plants) did not result in a rescue of the systemic spread of virus.
The HR is a general response of plants to attack by different pathogens, including Agrobacterium spp. Certain (incompatible) Agrobacterium spp–plant interactions lead to the development of PCD-like HR (Hansen, 2000). Although it remains to be determined whether the plant caspase-mediated fragmentation of VirD2 occurs during Agrobacterium-induced apoptosis, evidence supporting the involvement of caspase-like protease(s) in Agrobacterium-triggered PCD in maize cells has been reported (Hansen, 2000). It is tempting to speculate on a possible role of VirD2 fragmentation in the protection of plants from Agrobacterium-mediated transformation and tumor formation. Caspase cleavage detaches the NLS from VirD2, thus preventing its nuclear accumulation. Moreover, such a cleavage should abrogate the nuclear uptake of VirD2-linked T-DNA that is a prerequisite for the integration of T-DNA into the plant genome and hence its genetic transformation. Indeed, the presence of the functional NLS in VirD2 is important for tumor induction (Shurvinton et al., 1992), and removal of the C-terminal VirD2 region starting one amino acid residue downstream from the plant caspase cleavage site (identified in this work) abrogates tumorigenesis (Steck et al., 1990). Therefore, caspase activation should protect (defend) plants against agrobacterial transformation. It might be envisaged that, to achieve successful transformation, Agrobacterium has evolved a counterdefense system that disables caspase activation.
The challenge for the future is to purify to high homogeneity and characterize plant caspase and to identify a gene for this enzyme. This may provide new insights into the molecular mechanisms of PCD in plants and could lead to powerful approaches for the modulation of plant growth and development and for protection from pathogen attack and abiotic stresses. Moreover, comparative analysis of plant and animal caspases will illuminate elements of the PCD machinery that are conserved between the plant and animal kingdoms.
METHODS
Plasmid Construction
The protein-encoding portion of virD2 cDNA in pQE13 vector (Qiagen, Valencia, CA) was obtained from V. Lunin (Institute of Agricultural Biotechnology, Moscow). The virD2 cDNA then was excised from this plasmid as a 100-bp SauIIIA-PstI fragment and a 1240-bp PstI-XbaI DNA fragment and inserted between the BamHI and XbaI sites of pUC19 to produce pUC19/VirD2. Elimination of the Ecl136II-Bsp120I fragment from pUC19/VirD2, filling in with the Klenow fragment, and self-ligation of the rest of the plasmid produced pUC19/VirD2Ct.
Bacterial Expression
For overproduction of the full-length VirD2 protein with a (His)6 tag, pQE30/VirD2 was constructed by ligating a KpnI-SalI fragment from pUC19/VirD2 into pQE30 vector (Qiagen). To construct pQE32/VirD2Ct-GFP, an ApaI-BamHI fragment from pUC19/VirD2 and a NheI-NarI/Klenow fragment from pFRED25ΔNaegfphyg (Stauber et al., 1998; a gift from G. Pavlakis, National Cancer Institute, Frederick, MD) were inserted successively between the ApaI-BamHI and NheI-NotI/Klenow sites of pSL1180 (Amersham Pharmacia Biotech, Uppsala, Sweden), respectively, to produce pSL1180/VirD2Ct-GFP and then transferred, as a XmaI-PstI fragment, into similarly digested pQE32 (Qiagen).
Mutations were introduced in the virD2 sequence by PCR on pUC19/VirD2Ct with the mutagenic primers 5′-TCggaTcCACGCGGACGCTTTG-3′ (for VirD2CtΔNLS), 5′-CTACTGCCAgCCTGTTCGC-3′ [for VirD2Ct(D10A)], 5′-GACAATGAAgCGGTTGCG-3′ [for VirD2Ct(D39A)] (lowercase letters indicate the nucleotide substitutions; Figure 2A), and the pUC19 direct and reverse sequencing primers. The PCR products were digested with EcoRI and BamHI, ligated into pUC19, and sequenced to check for the absence of unwanted mutations. The double mutant D10,39A was constructed by DNA shuffling using a DdeI site located between the corresponding mutations. The mutated VirD2Ct genes were substituted for the wild-type gene in pSL1180/VirD2Ct-GFP using the Bsp120I and BamHI sites and then transferred into pQE32 vector as XmaI-PstI fragments.
To construct GFP-virD2Ct fusions, a Bsp120I/Klenow-XbaI DNA fragment from pUC19/VirD2 was inserted into pFRED25ΔNaegfphyg cut with NarI/Klenow and XbaI, yielding pFRED/GFP-VirD2Ct. A GFP-virD2Ct–encoding SacII/T4 polymerase-SacI fragment then was transferred into pSL1180 cut with KspAI and SacI to obtain pSL1180/GFP-VirD2Ct, and the HpaI-XbaI fragment from the latter plasmid was inserted into similarly cleaved pUC19 vector to produce pUC19/GFP-VirD2Ct. In this plasmid, the wild-type VirD2Ct-encoding Bsp120I-BamHI fragment was substituted by the analogous DNA fragment encoding VirD2CtΔNLS, yielding pUC19/GFP-VirD2CtΔNLS. Then, the Bsp120I-XbaI fragments encoding VirD2Ct with D/A mutations were substituted for the VirD2CtΔNLS gene (Figure 2A). Finally, the NheI/Klenow-HindIII fragments from the resulting series of pUC19/GFP-VirD2Ct derivatives were transferred into pQE31 vector (Qiagen) cut with BamHI/Klenow and HindIII to give pQE31/GFP-VirD2Ct variants. Note that the VirD2Ct and GFP domains are separated by 3– and 23–amino acid spacers in the GFP-VirD2Ct and VirD2Ct-GFP constructs, respectively.
The cDNA fragment encoding human caspase-3 was amplified by PCR from a HeLa cDNA library using primers 5′-CATGGAGAACACTGAAAAC-3′ and 5′-GGTGATAAAAATAGAGTTCTT-3′. An 830-bp PCR product was inserted into pBluescript II SK+ vector (Stratagene) between the SmaI and XhoI/Klenow sites, excised with NcoI (partial digestion) and XhoI, and inserted into the NcoI-XhoI–digested pET33b(+) vector (Novagen, Madison, WI). The structure of the resulting construct was confirmed by sequencing.
Expression in Plants
A Tobacco mosaic virus (TMV)–based vector, TMV(30B) (Shivprasad et al., 1999), was used for the production of chimeric proteins in tobacco (Nicotiana tabacum) plants. An ∼9.4-kb NcoI-XhoI fragment and an ∼300-bp NcoI-AgeI fragment obtained from p30B were ligated simultaneously with the AgeI-XhoI fragment from pSL1180/GFP-VirD2Ct encoding wild-type GFP-VirD2Ct or with the AgeI-SalI fragments from the pUC19/GFP-VirD2Ct series of plasmids encoding mutated VirD2Ct moiety (Figure 2A). The resulting plasmids were designated p30B/GFP-VirD2Ct, p30B/GFP-VirD2Ct(D10A), p30B/GFP-VirD2Ct(D39A), and p30B/GFP-VirD2Ct(D10,39A). To produce VirD2Ct-GFP in plants, the GFP-encoding NheI-XhoI fragment in pSL1180/VirD2Ct-GFP was substituted with an analogous fragment from pFRED25 (Stauber et al., 1998; a gift from G. Pavlakis), yielding pSL1180/VirD2Ct-GFPterm. The SmaI-XhoI fragment from this plasmid was inserted into pET33b(+) cut with NcoI/Klenow and XhoI, resulting in pET/ATGVirD2Ct-GFPterm, and then transferred, as a XbaI/Klenow-XhoI fragment, into HpaI- and XhoI-digested pSL1180. Finally, the AgeI-XhoI fragment from the resulting plasmid was ligated with the ∼9.4-kb NcoI-XhoI fragment and the ∼300-bp NcoI-AgeI fragment from p30B to produce p30B/VirD2Ct-GFP.
Purification of Recombinant Proteins
(His)6-tagged VirD2, GFP-VirD2Ct, and VirD2Ct-GFP were isolated from Escherichia coli JM109 cells freshly transformed with the respective plasmids. Cell cultures were grown in 25 mL of Luria-Bertani medium containing 100 μg/mL ampicillin at 37°C with shaking. Exponentially growing cells were induced with 0.5 mM isopropyl-β-d-thiogalactoside and harvested at 3 h after induction. Cell pellets were washed with TBST buffer (20 mM Tris-HCl, pH 7.5, 140 mM NaCl, and 0.05% Tween 20), resuspended in 1 mL of buffer A (20 mM Tris-HCl, pH 8.0, 50 mM NaCl, and 5% glycerol) supplemented with aprotenin (4 μg/mL), leupeptin (2 μg/mL), and phenylmethylsulfonyl fluoride (PMSF; 1.2 mM), sonicated on ice by four 30-s bursts, and centrifuged at 12,000g for 10 min. The supernatant was discarded, and the pellet was resuspended in 1 mL of buffer B (10 mM Tris-HCl, pH 8.0, 1 M NaCl, and 8 M urea), incubated for 30 min at room temperature, and sonicated on ice by three 30-s bursts. After centrifugation at 12,000g for 10 min, cleared lysates were incubated in batches with nickel–nitrilotriacetic acid (Ni-NTA) agarose (Qiagen) equilibrated with buffer B for 3 h at room temperature with shaking. The resin then was washed with 5 × 1 mL of buffer C (10 mM Tris-HCl, pH 6.3, 100 mM NaCl, 100 mM Na2HPO4, and 8 M urea), and the proteins were eluted with 400 μL of buffer C containing 100 mM EDTA for 1 h at room temperature with shaking and then with two additional 100-μL portions of the same buffer. The eluates were combined and dialyzed overnight against 200 mL of buffer 20/100 (20 mM Hepes, pH 6.8, 100 mM NaCl, 2 mM DTT, and 0.05% Tween 20) or against B1 buffer, pH 5.5 (50 mM Hepes, pH 5.5, 50 mM NaCl, 2 mM DTT, 0.1% Tween 20, and 5% glycerol).
E. coli BL21(DE3) cells freshly transformed with a caspase-3–encoding plasmid were grown in 80 mL of Luria-Bertani medium containing 25 μg/mL kanamycin at 30°C with shaking. Exponentially growing cells were induced with 0.5 mM isopropyl-β-d-thiogalactoside and harvested at 3.5 h after induction. The cell pellet was resuspended in 8 mL of buffer SB (50 mM Hepes, pH 6.8, and 100 mM NaCl) containing 2 μg/mL aprotenin and 1 mM PMSF and sonicated on ice by four 30-s bursts. After centrifugation, the precipitate of cell debris was discarded and the supernatant was incubated with shaking with 300 μL of Ni-NTA agarose at 0°C for 3 h. The resin then was washed stepwise with SB buffer containing 25 mM imidazole (5 × 1 mL), and caspase-3 was eluted with two 250-μL portions of a buffer containing 50 mM Hepes, 100 mM NaCl, and 250 mM imidazole, pH 6.3. The combined eluates were dialyzed overnight against 200 mL of a buffer containing 50 mM Hepes, pH 6.8, 100 mM NaCl, 2 mM DTT, and 5% glycerol and stored at −70°C. Alternatively, a portion of the caspase-3 sample was dialyzed against B1 buffer, pH 5.5.
In Vitro Transcription, Inoculation of Plants, Confocal Laser Scanning Microscopy, and Fluorescence Microscopy
Plasmids were linearized by digestion with KpnI, and in vitro transcripts were synthesized with T7 RNA polymerase using the mMESSAGE mMACHINE T7 Kit (Ambion, Austin, TX). The transcripts derived from 0.2 μg of plasmid template were inoculated directly to corundum-dusted leaves of tobacco cv Samsun NN, Xanthi nc, or Samsun plants. Inoculated plants were maintained in a growth cabinet at 33°C with a 16-h photoperiod for 48 to 60 h before transfer to a similar cabinet at 24°C to induce and synchronize the hypersensitive response.
Infected leaves were viewed using a stereofluorescence microscope. To visualize nuclear DNA, leaf tissues were stained with 4′,6-diamidino-2-phenylindole (1 μg/mL). Fluorescent infection sites also were monitored using an MRC 1000 confocal laser scanning microscope (Bio-Rad, Hercules, CA) equipped with a 25-mW krypton/argon laser. For GFP imaging, blue excitation at 488 nm with emission filter 522 DF (32 nm) was used.
For experiments with synthetic caspase inhibitors, potential inhibitors, biotinyl-TATD-CHO (custom synthesized by Bachem, Merseyside, UK), or biotinyl-DEVD-CHO (Bachem) was dissolved in DMSO, diluted in 0.01 M phosphate buffer, pH 5.5, to a final concentration of 2 mM, mixed with an equal volume of purified TMV preparation, and inoculated immediately into tobacco cv Samsun NN leaves as described above. The same volume of DMSO was added to control TMV samples. Infected plants were kept at 24°C.
Protein Gel Blot Analysis
For immunoblot analysis, samples of plant tissues from fluorescent infection sites (detected by illuminating with long-wavelength UV light) were ground in liquid nitrogen, resuspended in dissociation buffer (50 mM Tris-HCl, pH 6.8, containing 2 M urea, 3% SDS, and 1.5% β-mercaptoethanol), and incubated at 95°C for 5 min. Samples then were separated by electrophoresis on 10% SDS-polyacrylamide gels using the Tris-Tricine system (Schagger and von Jagow, 1987). Proteins were transferred to a nitrocellulose membrane (Protran BA85; Schleicher & Schuell, Dietzenbach, Germany), and blots were treated with the polyclonal antibody prepared against GFP. Immunoreactions were detected using the enhanced chemiluminescence protein gel blot system (Amersham Pharmacia Biotech).
Partial Purification of Tobacco Caspase
Fresh tobacco Xanthi nc (carrying the N gene) leaves (7 g) were infected with the wild-type TMV (U-1 strain), kept at 33°C for 2 days, and then transferred to 24°C until multiple necrotic lesions became clearly visible (usually 2 to 5 days). The leaves were ground under liquid nitrogen, and the sample was suspended in 25 mL of extraction buffer B1, pH 6.8, lacking NaCl and containing 1 mM PMSF and sonicated for 10 min. The debris were eliminated by two successive 15-min centrifugation steps at 15,000g, the supernatant was desalted by passing through a Sephadex G-25 column (2.8 × 17 cm) equilibrated with the same buffer, and the flow-through fractions were collected. After diluting with the buffer up to 85 mL, the sample was loaded onto a DE52 column (volume = 8.5 mL), the column was washed with B1 buffer, pH 6.8, lacking NaCl, and the proteins were eluted with the same buffer containing 0 to 0.2 M NaCl. Protein fractions were monitored for plant caspase activity using recombinant GFP-VirD2Ct as a substrate and 12% SDS-PAGE for fractionation of the reaction mixtures. Proteins eluted at ∼70 mM NaCl and displaying putative caspase activity were combined, diluted up to 50 mL with B1 buffer, pH 7.8, lacking NaCl, and applied to a 3-mL DE52 column equilibrated with the same buffer. The column was washed with the buffer, and the proteins were eluted with the same buffer containing 0 to 0.2 M NaCl. The peak 0.5-mL fraction eluted at 100 mM NaCl was dialyzed against B1 buffer, pH 5.5, and stored at −70°C.
Caspase Hydrolysis and Inhibition
Purified VirD2 derivatives, in 3-μg aliquots, were treated with 0.15 μg of recombinant human caspase-3 in a total volume of 10 μL of 20/100 buffer at 37°C for 1 h. Where indicated, caspase-3 was preincubated with 10 μM Ac-DEVD-CHO (Bachem) (from stock solution in DMSO) at 27°C for 20 min before its addition to the substrate. An equivalent amount of DMSO was added to the sample lacking the inhibitor. To assess the presence of the (His6) tag sequence in the cleavage products of full-length VirD2, dry urea was added to the reaction mixture up to 8 M and the sample was loaded onto 20 μL of Ni-NTA agarose equilibrated with buffer B for 1 h at room temperature. The flow-through was collected after brief centrifugation, the resin was washed with the loading buffer, and the bound proteins were eluted from the affinity resin by boiling in Laemmli (1970) sample buffer.
Treatment of 3-μg aliquots of GFP-VirD2Ct with partially purified plant caspase (1.0 μL of the enzyme preparation per sample) was performed in B1 buffer, pH 5.5, in a total volume of 15 μL at 27°C for 1.5 h. For direct comparison of the inactivation of human and plant enzymes with inhibitors, caspase-3 hydrolysis and inhibition, as depicted in Figures 6B and 6D, also were performed in B1 buffer, pH 5.5. Enzymes were preincubated in the same buffer with the indicated concentrations of zVAD-fmk, biotinyl-DEVD-CHO, or biotinyl-TATD-CHO at 27°C for 1 h before their addition to the substrate. Alternatively, enzymes were preincubated for 30 min at 27°C (plant caspase) or 37°C (caspase-3) in B1 buffer, pH 5.5, containing 200 μM DTT in the presence of 1 mM 4-(2-aminoethyl) benzenesulfonyl fluoride, 100 μM chymostatin, 10 μM E-64, 5 mM N-ethylmaleimide, 200 μM HgCl2, 5 mM EDTA, or 1 μM pepstatin. Then, the substrate was added and the samples were incubated at the temperatures indicated for 2 h. Reaction mixtures were fractionated by 12% SDS-PAGE, and proteins were stained with Coomassie Brilliant Blue R 250.
Recombinant human prothymosin α (a gift from A. Evstafieva, Belozersky Institute, Moscow), in 2-μg aliquots, was treated with 0.15 μg of caspase-3 or with a 2.0-μL aliquot of the partially purified plant caspase. Hydrolysis was performed in a total volume of 10 μL of B1 buffer, pH 5.5, at 37°C for 2 h. The reaction products were precipitated with ethanol and analyzed with 8% PAGE and 7 M urea as described (Evstafieva et al., 2003).
Analysis of TMV Multiplication
For ELISA analyses of TMV multiplication in infected leaves, samples of plant tissues (0.2 g) from inoculated leaves were homogenized in 1 mL of PBS, pH 7.2, containing 0.05% Tween 20. The double antibody sandwich ELISA procedure (Clark et al., 1986) was used to detect and quantify TMV antigen using polyclonal TMV antiserum.
RNA Isolation and Reverse Transcriptase–Mediated PCR Analysis
Total RNA was extracted from plants using RNAwiz solution (Ambion) and treated with RNase-free DNase I (Ambion). Reverse transcriptase–mediated PCR analysis of HSR203J and PR-1a gene expression was performed using the following oligonucleotide primer combinations (for HSR203J, 5′-ATGGTTCATGAAAAGCA-3′ as a forward primer and 5′-CGGTGCTCCGGCGCGAG-3′ as a reverse primer; for PR-1a, 5′-ATGGGATTTGTTCTCT-3′ as a forward primer and 5′-AGCCTTAGCAGCCGTCA-3′ as a reverse primer). First-strand cDNA was synthesized using 5 μg of total RNA, oligo(dT) primer, and SuperScript RNase H− reverse transcriptase (Invitrogen Life Technologies, Paisley, UK) according to the manufacturer's recommendations. Ubiquitin mRNA, used as a control, was generated using 5′-ATGCAGATCTTCGTGA-3′ as a forward primer and 5′-TAGTCAGCCAAGGTCCT-3′ as a reverse primer. First-strand cDNA was used as a template for PCR product amplification through 25, 30, 35, 40, 45, and 50 cycles. The intensities of PCR-generated fragments were analyzed and quantified using the Intelligent Quantifier, version 2.5.0 (BioImage, Ann Arbor, MI).
Mass Spectrometric Analysis of Caspase Cleavage Sites in GFP-VirD2Ct
Bands excised from imidazole-zink-negative stained gels containing either GFP-VirD2Ct or its cleavage products from capase-3 or tobacco caspase treatment (Figure 5) were subjected to endoproteinase LysC digestion essentially as described by Evstafieva et al. (2003). Gel-contained proteins were reduced in the presence of 10 mM tris(carboxyethyl)phosphine and alkylated using 50 mM iodoacetamide. To remove remaining reagents, the gel pieces were shrunk subsequently in acetonitrile, rehydrated in 25 mM Tris-HCl and 1 mM EDTA, pH 8.5, and dehydrated again. A second rehydration step was performed in the same buffer plus 150 ng per sample of LysC endoproteinase (Roche Diagnostics, Indianapolis, IN). After overnight incubation at 37°C, samples were acidified with 15 μL of 0.5% trifluoroacetic acid (TFA) and mixed for 4 h in the presence of a minute amount of conditioned Poros R20 reversed-phase chromatography beads (Applied Biosystems, Foster City, CA). The beads were collected on ZipTips (Millipore, Bedford, MA) that were allowed to function as a collecting frit for the Poros material. After washing with 10 μL of 0.1% TFA, 3 μL of elution solvent consisting of 50% methanol, 20% acetonitrile, and 0.1% TFA was applied to elute the peptides directly onto a matrix-assisted laser-desorption ionization (MALDI) sample plate. The sample drops were mixed with the MALDI matrix—0.5 μL of one-fourth–saturated α-cyano-4-hydroxy cinnamic acid in the same solvent—and allowed to dry.
Spectra were acquired on a Voyager-DE STR MALDI mass spectrometer (ABI) in linear mode. Singly charged ions of bovine insulin and ubiquitin served as external calibration standards using average masses of the corresponding singly charged protonated species. The spectra were recalibrated internally on peaks identified as LysC digestion products of GFP-VirD2Ct at m/z 1534.76 and 4732.15.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact M.E. Taliansky, mtalia@scri.sari.ac.uk; or A.B. Vartapetian, varta@genebee.msu.su.
Accession Number
The accession number for the virD2 cDNA is NP536300.
Acknowledgments
We thank V. Lunin for providing VirD2 cDNA, G. Pavlakis for providing vectors encoding a bright version of GFP, A. Evstafieva for providing human prothymosin α, T. Pestova and C. Hellen for their constant support, and S. MacFarlane and B. Harrison for critical reading of the manuscript. This work was supported in part by the Ludwig Institute for Cancer Research (to A.B.V.), by a grant-in-aid from the Scottish Executive Environment and Rural Affairs Department (to M.E.T.), and by grants from the Russian Foundation for Basic Research (to N.V.C. and A.B.V.), the U.S. Civilian Research and Development Foundation for the Independent States of the Former Soviet Union (to A.B.V.), the Russian Frontiers in Genetics Program (to A.B.V.), and the Royal Society (ex-quota visit fellowship to N.O.K.).
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.017889.
References
- Adrain, C., and Martin, S.J. (2001). The mitochondrial apoptosome: A killer unleashed by the cytochrome seas. Trends Biochem. Sci. 26, 390–397. [DOI] [PubMed] [Google Scholar]
- Balk, J., and Leaver, C.J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen, H.M., Zhou, J., and Dai, Y.R. (2000). Cleavage of lamin-like proteins in in vivo and in vitro apoptosis of tobacco protoplasts induced by heat shock. FEBS Lett. 480, 165–168. [DOI] [PubMed] [Google Scholar]
- Clark, M.F., Lister, R.M., and Bar-Joseph, M. (1986). ELISA techniques. Methods Enzymol. 118, 742–766. [Google Scholar]
- Clarke, A., Desikan, R., Hurst, R.D., Hancock, J.T., and Neill, S.J. (2000). NO way back: Nitric oxide and programmed cell death in Arabidopsis thaliana suspension cultures. Plant J. 24, 667–677. [DOI] [PubMed] [Google Scholar]
- Danon, A., Delorme, V., Mailhac, N., and Gallois, P. (2000). Plant programmed cell death: A common way to die. Plant Physiol. Biochem. 38, 647–655. [Google Scholar]
- del Pozo, O., and Lam, E. (1998). Caspases and programmed cell death in the hypersensitive response of plants to pathogens. Curr. Biol. 8, 1129–1132. [DOI] [PubMed] [Google Scholar]
- del Pozo, O., and Lam, E. (2003). Expression of the baculovirus p35 protein in tobacco affects cell death progression and compromises N gene-mediated disease resistance response to Tobacco mosaic virus. Mol. Plant-Microbe Interact. 16, 485–494. [DOI] [PubMed] [Google Scholar]
- Dickman, M.B., Park, Y.K., Oltersdorf, T., Li, W., Clemente, T., and French, R. (2001). Abrogation of disease development in plants expressing animal antiapoptotic genes. Proc. Natl. Acad. Sci. USA 98, 6957–6962. [DOI] [PMC free article] [PubMed] [Google Scholar]
- D'Silva, I., Poirier, G.G., and Heath, M.C. (1998). Activation of cysteine proteases in cowpea plants during the hypersensitive response: A form of programmed cell death. Exp. Cell Res. 245, 389–399. [DOI] [PubMed] [Google Scholar]
- Durrenberger, F., Crameri, A., Hohn, B., and Koukolikova-Nicola, Z. (1989). Covalently bound VirD2 protein of Agrobacterium tumefaciens protects the T-DNA from exonucleolytic degradation. Proc. Natl. Acad. Sci. USA 86, 9154–9158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ekert, P.G., Silke, J., and Vaux, D.L. (1999). Caspase inhibitors. Cell Death Differ. 6, 1081–1086. [DOI] [PubMed] [Google Scholar]
- Elbaz, M., Avni, A., and Weil, M. (2002). Constitutive caspase-like machinery executes programmed cell death in plant cells. Cell Death Differ. 9, 726–733. [DOI] [PubMed] [Google Scholar]
- Evstafieva, A.G., Belov, G.A., Rubtsov, Y.P., Kalkum, M., Joseph, B., Chichkova, N.V., Sukhacheva, E.A., Bogdanov, A.A., Pettersson, R.F., Agol, V.I., and Vartapetian, A.B. (2003). Apoptosis-related fragmentation, translocation, and properties of human prothymosin alpha. Exp. Cell Res. 284, 209–221. [DOI] [PubMed] [Google Scholar]
- Green, D.R. (1998). Apoptotic pathways: The roads to ruin. Cell 94, 695–698. [DOI] [PubMed] [Google Scholar]
- Greenberg, J.T. (1996). Programmed cell death: A way of life for plants. Proc. Natl. Acad. Sci. USA 93, 12094–12097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenberg, J.T. (1997). Programmed cell death in plant-pathogen interactions. Annu. Rev. Plant Physiol. Plant Mol. Biol. 48, 525–545. [DOI] [PubMed] [Google Scholar]
- Hansen, G. (2000). Evidence for Agrobacterium-induced apoptosis in maize cells. Mol. Plant-Microbe Interact. 13, 649–657. [DOI] [PubMed] [Google Scholar]
- Heath, M.C. (2000). Hypersensitive response-related death. Plant Mol. Biol. 44, 321–334. [DOI] [PubMed] [Google Scholar]
- Hoeberichts, F.A., and Woltering, E.J. (2003). Multiple mediators of plant programmed cell death: Interplay of conserved cell death mechanisms and plant-specific regulators. Bioessays 25, 47–57. [DOI] [PubMed] [Google Scholar]
- Howard, E.A., Zupan, J.R., Citovsky, V., and Zambryski, P.C. (1992). The VirD2 protein of A. tumefaciens contains a C-terminal bipartite nuclear localization signal: Implications for nuclear uptake of DNA in plant cells. Cell 68, 109–118. [DOI] [PubMed] [Google Scholar]
- Kassanis, B. (1952). Some effects of high temperature on susceptibility of plants to infection with viruses. Ann. Appl. Biol. 26, 358–369. [Google Scholar]
- Kaufmann, S.H., and Hengartner, M.O. (2001). Programmed cell death: Alive and well in the new millennium. Trends Cell Biol. 11, 526–534. [DOI] [PubMed] [Google Scholar]
- Kerr, J.F., Wyllie, A.H., and Currie, A.R. (1972). Apoptosis: A basic biological phenomenon with wide-ranging implications in tissue kinetics. Br. J. Cancer 26, 239–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacomme, C., and Santa Cruz, S. (1999). Bax-induced cell death in tobacco is similar to the hypersensitive response. Proc. Natl. Acad. Sci. USA 96, 7956–7961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laemmli, U.K. (1970). Cleavage of structural proteins during the assembly of the head of bacteriophage T4. Nature 227, 680–685. [DOI] [PubMed] [Google Scholar]
- Lam, E., and del Pozo, O. (2000). Caspase-like protease involvement in the control of plant cell death. Plant Mol. Biol. 44, 417–428. [DOI] [PubMed] [Google Scholar]
- Lam, E., Kato, N., and Lawton, M. (2001). Programmed cell death, mitochondria and the plant hypersensitive response. Nature 411, 848–853. [DOI] [PubMed] [Google Scholar]
- Minami, A., and Fukuda, H. (1995). Transient and specific expression of a cysteine endopeptidase associated with autolysis during differentiation of Zinnia mesophyll cells into tracheary elements. Plant Cell Physiol. 36, 1599–1606. [PubMed] [Google Scholar]
- Mitsuhar, I., Malik, K.A., Miura, M., and Ohashi, Y. (1999). Animal cell death suppressors Bcl-x(L) and Ced-9 inhibit cell death in tobacco plants. Curr. Biol. 9, 775–778. [DOI] [PubMed] [Google Scholar]
- Mlejnek, P., and Prochazka, S. (2002). Activation of caspase-like proteases and induction of apoptosis by isopentenyladenosine in tobacco BY-2 cells. Planta 215, 158–166. [DOI] [PubMed] [Google Scholar]
- Pontier, D., Godiard, L., Marco, Y., and Roby, D. (1994). hsr203J, a tobacco gene whose activation is rapid, highly localized and specific for incompatible plant pathogen interactions. Plant J. 5, 507–521. [DOI] [PubMed] [Google Scholar]
- Sanchez, P., de Torres Zabala, M., and Grant, M. (2000). AtBI-1, a plant homologue of Bax inhibitor-1, suppresses Bax-induced cell death in yeast and is rapidly upregulated during wounding and pathogen challenge. Plant J. 21, 393–399. [DOI] [PubMed] [Google Scholar]
- Schagger, H., and von Jagow, G. (1987). Tricine-sodium dodecyl sulfate-polyacrylamide gel electrophoresis for the separation of proteins in the range from 1 to 100 kDa. Anal. Biochem. 166, 368–379. [DOI] [PubMed] [Google Scholar]
- Schmid, M., Simpson, D., and Gietl, C. (1999). Programmed cell death in castor bean endosperm is associated with the accumulation and release of a cysteine endopeptidase from ricinosomes. Proc. Natl. Acad. Sci. USA 96, 14159–14164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shivprasad, S., Pogue, G.P., Lewandowski, D.J., Hidalgo, J., Donson, J., Grill, L.K., and Dawson, W.O. (1999). Heterologous sequences greatly affect foreign gene expression in tobacco mosaic virus-based vectors. Virology 255, 312–323. [DOI] [PubMed] [Google Scholar]
- Shurvinton, C.E., Hodges, L., and Ream, W. (1992). A nuclear localization signal and the C-terminal omega sequence in the Agrobacterium tumefaciens VirD2 endonuclease are important for tumor formation. Proc. Natl. Acad. Sci. USA 89, 11837–11841. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Solomon, M., Belenghi, B., Delledonne, M., Menachem, E., and Levine, A. (1999). The involvement of cysteine proteases and protease inhibitor genes in the regulation of programmed cell death in plants. Plant Cell 11, 431–444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stauber, R.H., Carney, P., Horie, K., Hudson, E.A., Tarasova, N.I., Gaitanaris, G.A., and Pavlakis, G.N. (1998). Development and applications of enhanced autofluorescent protein mutants. Biotechniques 24, 462–471. [DOI] [PubMed] [Google Scholar]
- Steck, T.R., Lin, T.S., and Kado, C.I. (1990). VirD2 gene product from the nopaline plasmid pTiC58 has at least two activities required for virulence. Nucleic Acids Res. 18, 6953–6958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sun, Y.L., Zhao, Y., Hong, X., and Zhai, Z.H. (1999). Cytochrome c release and caspase activation during menadione-induced apoptosis in plants. FEBS Lett. 462, 317–321. [DOI] [PubMed] [Google Scholar]
- Talanian, R.V., Quinlan, C., Trautz, S., Hackett, M.C., Mankovich, J.A., Banach, D., Ghayur, T., Brady, K.D., and Wong, W.W. (1997). Substrate specificities of caspase family proteases. J. Biol. Chem. 272, 9677–9682. [DOI] [PubMed] [Google Scholar]
- Thornberry, N.A., and Lazebnik, Y. (1998). Caspases: Enemies within. Science 281, 1312–1316. [DOI] [PubMed] [Google Scholar]
- Tian, R., Zhang, G.Y., Yan, C.H., and Dai, Y.R. (2000). Involvement of poly(ADP-ribose) polymerase and activation of caspase-3-like protease in heat shock-induced apoptosis in tobacco suspension cells. FEBS Lett. 474, 11–15. [DOI] [PubMed] [Google Scholar]
- Tinland, B., Schoumacher, F., Gloeckler, V., Bravo-Angel, A.M., and Hohn, B. (1995). The Agrobacterium tumefaciens virulence D2 protein is responsible for precise integration of T-DNA into the plant genome. EMBO J. 14, 3585–3595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Uren, A.G., O'Rourke, K., Aravind, L.A., Pisabarro, M.T., Seshagiri, S., Koonin, E.V., and Dixit, V.M. (2000). Identification of paracaspases and metacaspases: Two ancient families of caspase-like proteins, one of which plays a key role in MALT lymphoma. Mol. Cell 6, 961–967. [DOI] [PubMed] [Google Scholar]
- Ward, E.R., and Barnes, W.M. (1988). VirD2 protein of Agrobacterium tumefaciens very tightly linked to the 5′ end of T-strand DNA. Science 242, 927–930. [Google Scholar]
- Wolf, B.B., and Green, D.R. (1999). Suicidal tendencies: Apoptotic cell death by caspase family proteinases. J. Biol. Chem. 274, 20049–20052. [DOI] [PubMed] [Google Scholar]
- Yanofsky, M.F., Porter, S.G., Young, C., Albright, L.M., Gordon, M.P., and Nester, E.W. (1986). The virD operon of Agrobacterium tumefaciens encodes a site-specific endonuclease. Cell 47, 471–477. [DOI] [PubMed] [Google Scholar]
- Zheng, T.S., Hunot, S., Kuida, K., and Flavell, R.A. (1999). Caspase knockouts: Matters of life and death. Cell Death Differ. 6, 1043–1053. [DOI] [PubMed] [Google Scholar]
- Ziemienowicz, A., Merkle, T., Schoumacher, F., Hohn, B., and Rossi, L. (2001). Import of Agrobacterium T-DNA into plant nuclei: Two distinct functions of VirD2 and VirE2 proteins. Plant Cell 13, 369–384. [DOI] [PMC free article] [PubMed] [Google Scholar]