Abstract
AIMS
Little information is available regarding the metabolic routes of anastrozole and the specific enzymes involved. We characterized anastrozole oxidative and conjugation metabolism in vitro and in vivo.
METHODS
A sensitive LC-MS/MS method was developed to measure anastrozole and its metabolites in vitro and in vivo. Anastrozole metabolism was characterized using human liver microsomes (HLMs), expressed cytochrome P450s (CYPs) and UDP-glucuronosyltransferases (UGTs).
RESULTS
Hydroxyanastrozole and anastrozole glucuronide were identified as the main oxidative and conjugated metabolites of anastrozole in vitro, respectively. Formation of hydroxyanastrozole from anastrozole was markedly inhibited by CYP3A selective chemical inhibitors (by >90%) and significantly correlated with CYP3A activity in a panel of HLMs (r = 0.96, P = 0.0005) and mainly catalyzed by expressed CYP3A4 and CYP3A5. The Km values obtained from HLMs were also close to those from CYP3A4 and CYP3A5. Formation of anastrozole glucuronide in a bank of HLMs was correlated strongly with imipramine N-glucuronide, a marker of UGT1A4 (r = 0.72, P < 0.0001), while expressed UGT1A4 catalyzed its formation at the highest rate. Hydroxyanastrozole (mainly as a glucuronide) and anastrozole were quantified in plasma of breast cancer patients taking anastrozole (1 mg day−1); anastrozole glucuronide was less apparent.
CONCLUSION
Anastrozole is oxidized to hydroxyanastrozole mainly by CYP3A4 (and to some extent by CYP3A5 and CYP2C8). Once formed, this metabolite undergoes glucuronidation. Variable activity of CYP3A4 (and probably UGT1A4), possibly due to genetic polymorphisms and drug interactions, may alter anastrozole disposition and its effects in vivo.
Keywords: anastrozole, breast cancer, glucuronidation, hydroxylation, in vitro, in vivo
WHAT IS ALREADY KNOWN ABOUT THIS SUBJECT
Anastrozole is primarily cleared by hepatic metabolism via oxidative and conjugating enzymes.
WHAT THIS STUDY ADDS
Anastrozole is oxidized to hydroxyanastrozole mainly by CYP3A4/5 and glucuronidated to anastrozole glucuronide predominantly by UGT1A4 in vitro.
Hydroxyanastrozole glucuronide and hydroxyanastrozole were quantified as the major metabolites of anastrozole in plasma of breast cancer patients.
This study describes for the first time anastrozole metabolic pathways and the enzymes involved, which may serve as the scientific basis for pharmacogenetic and drug-interaction assessments.
Introduction
Globally, breast cancer is the most common cancer affecting women and one of the leading causes of death [1]. In the US, one woman in eight will develop the disease during the course of her life [2]. Approximately 80% of breast cancers in post-menopausal women are oestrogen receptor-positive [3]. Since these tumours have cell-surface oestrogen receptors that bind with oestrogens, leading to cancer cell proliferation, depletion of circulating and tissue concentrations of oestrogen or blockade of the cellular effects of oestrogen by binding to oestrogen receptors is an effective strategy to reduce tumour growth and slow disease progression [3, 4].
For over 30 years, tamoxifen has been at the forefront for the treatment and prevention of oestrogen receptor-positive (ER-positive) breast cancers in pre- and post-menopausal women [3]. More recently, potent and selective inhibitors of aromatase [cytochrome P-450 (CYP) 19], a rate-limiting enzyme in the biosynthesis of oestrogens have been effectively used to deplete plasma and tissue concentrations of oestrogens in post-menopausal women [4]. Currently, three drugs (letrozole, anastrozole and exemestane) that belong to the third generation aromatase inhibitors are commonly prescribed to treat postmenopausal women with breast cancer. The clinical use of aromatase inhibitors has been on the rise in recent years because large randomized clinical trials have shown that aromatase inhibitors are superior to tamoxifen in adjuvant therapy [5–7].
Anastrozole, [2,2′-[5-1H-1,2,4-triazole-1-y-methyl)-1,3-phenylene]bis(2-methylpropiononitrile)] (Figure 1), belongs to the non-steroidal triazole-derivative group of aromatase inhibitors. On the basis of favourable efficacy and toxicity findings of anastrozole in the Arimidex, Tamoxifen Alone or in Combination trial (ATAC) compared with tamoxifen [5], anastrozole was approved by the US Food and Drug Administration in 2002 for adjuvant treatment of postmenopausal, early-stage hormone-sensitive breast cancer and is currently regarded as the first-line therapy for this indication. In addition, in the ATAC trial, there was a further reduction of 44% in the incidence of new contralateral breast cancers in the hormone-receptor positive women assigned to the anastrozole group compared with the tamoxifen group. This has stimulated interest in aromatase inhibitors as potential chemoprevention agents and is the basis for an ongoing large multicentre trial comparing the two drugs for prevention of a new primary breast cancer in women with a history of hormone-receptor positive ductal carcinoma in situ (DCIS) or early stage breast cancer [5].
Figure 1.
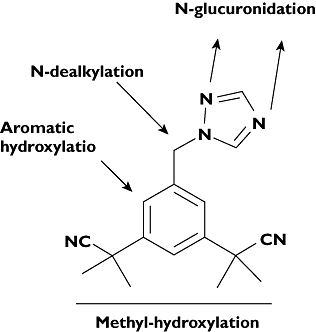
Anastrozole chemical structure and potential sites of metabolism
However, emerging data suggest high interindividual variability in the beneficial and adverse effects that include musculoskeletal disorders and joint pains and bone fracture to anastrozole and other aromatase inhibitor drugs [8]. Anastrozole is predominantly cleared by hepatic metabolism and evidence exists that its pharmacokinetics vary widely among patients [9]. Therefore, it is conceivable that differences in anastrozole metabolism may contribute to the overall variability experienced by patients.
Approximately 60% of the dose administered was excreted as metabolites and ∼10% was excreted in urine as unchanged after administration of a single radiolabelled dose of 1mg anastrozole to humans [8]. However, detailed information on how this drug is metabolized is lacking. Information provided in the product label of the drug state that triazole, anastrozole glucuronide and hydroxyanastrozole glucuronide have been identified in human urine and plasma, suggesting involvement of P450s (N-dealkylation and hydroxylation) and UGTs (glucuronidation) [8]. Recently, Ingle et al. reported that hydroxyanastrozole is the main metabolite of anastrozole in patients [9]. In a review article citing unpublished data, Lazarus & Sun [10] report that anastrozole undergoes direct N-glucuronidation by UGT1A4. The potential sites of metabolism are shown in Figure 1. However, there are no published data that confirm these findings or provide information about the quantitative contribution of each metabolic pathway to the overall clearance of the drug. More importantly, the specific enzymes responsible for anastrozole oxidative metabolism remain unknown, making prediction of factors controlling its pharmacokinetics difficult. In one in vitro study, anastrozole has been shown to inhibit competitively CYP1A2, CYP2C8/9 and CYP3A4 [2], but there are no published data that systematically address the contribution of these or other enzymes in vitro or in vivo.
The main purposes of the present study were (i) to develop a sensitive and reliable LC-MS/MS method to measure anastrozole and its metabolites in vitro and in vivo, (ii) to characterize anastrozole metabolic pathways and identify enzymes catalyzing them by conducting a comprehensive series of studies in human liver microsomal preparations, expressed CYPs and UGTs and (iii) to determine plasma steady state primary and secondary metabolism of anastrozole in breast cancer patients treated with 1 mg day−1 anastrozole.
Methods
Chemicals
Anastrozole (for preclinical evaluation) was obtained from Toronto Research Chemicals Inc. (Toronto, Canada). Desmethyldiazepam, diethyldithiocarbamate, ketoconazole, furafylline, omeprazole, pilocarpine, quercetin, quinidine, thioTEPA, ticlopidine, troleandomycin (oleandomycin triacetate), glucose 6-phosphate, glucose-6-phosphate dehydrogenase, NADP, alamethicin, magnesium chloride, β-glucuronidase (from Escherichia coli), Tris-HCl, UDPGA, imprimine, p-nitrophenol and testosterone were purchased from Sigma-Aldrich (St Louis, MO, USA). Sulfaphenazole was obtained from Ultrafine Chemicals (Manchester, UK). Rabbit antihuman UGT1A primary antibody was purchased from Santa Cruz Biotechnology, Inc. (Santa Cruz, California, USA). Rabbit antihuman UGT2B7 primary and secondary antibodies were purchased from BD Gentest (Woburn, MA, USA). Precast 10% acrylamide gels were obtained from Bio-Rad Laboratories (Hercules, California, USA). Blotto was purchased from Pierce Biotechnology (Rockford, IL, USA). Chemiluminescence reagents were obtained from GE Healthcare (Piscataway, NJ, USA). All other reagents were of HPLC grade or of the highest grade commercially available.
Human liver microsomes (HLMs)
Some HLMs that we used were either prepared from human liver tissues that were medically unsuitable for transplantation or were purchased from BD Gentest (Woburn, MA) and Cellz Direct (Austin, TX). Microsomal fractions were prepared by ultracentrifugation using standard protocols. Protein concentrations were determined by the Bradford method [11], using bovine serum albumin as a standard. The microsomal pellets were suspended in a reaction buffer to a protein concentration of 10 mg ml−1 (stock) and were kept at −80°C until used. Baculovirus-insect cell-expressed human P450s (1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4, and 3A5) (with oxidoreductase and cytochrome b5) and UGTs (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17) were purchased from BD Gentest Corp. (Woburn, MA, USA). Other HLMs obtained from 53 Caucasian donors were processed through Dr Mary Relling's laboratory at St Jude Children's Research Hospital (Memphis, TN, USA) and were provided by the Liver Tissue Procurement and Distribution System (funded by #NO1-DK-9-2310) and by the Cooperative Human Tissue Network.
A. Anastrozole oxidation in vitro
General incubation conditions
Incubation experiments were performed in HLMs to identify oxidative metabolites and to optimize conditions for incubation and LC-MS/MS analysis. Anastrozole was dissolved and serially diluted in methanol to the required concentration. Methanol was removed by drying in a speed vacuum before reconstituting the residue with phosphate buffer and addition of other incubation components. For the pilot studies, anastrozole (10 µm) and a NADPH-generating system (13 mm NADP, 33 mm glucose 6-phosphate, 33 mm MgCl2, and 0.4 U ml−1 glucose 6-phosphate dehydrogenase) in potassium phosphate buffer (pH 7.4) were allowed to equilibrate for 5 min at 37°C. The reaction was initiated by adding HLMs (0.5 mg protein ml−1) and allowed to proceed for 30 min at 37°C (final volume, 250 µl). Then, the reaction was terminated by placing tubes on ice and immediately adding 500 µl of acetonitrile. Desmethyldiazepam (20 µl of 250 ng ml−1) was added as an internal standard to the incubation sample which was then vortex mixed for 30 s, and centrifuged at 14 000 rev min−1 for 5 min in an Eppendorf model 5415D centrifuge (Brinkmann Instruments, Westbury, NY). The supernatant was extracted with ethyl acetate under alkaline pH (0.5 ml of 0.5 m NaOH/glycine buffer, adjusted to pH 10). After the sample was centrifuged for 15 min at 2500 rev min−1 in a Beckman GS-6R centrifuge (Global Medical Instrumentation, Inc., Ramsey, Minnesota), the organic phase was removed and evaporated to dryness. The residue was reconstituted with mobile phase from which an aliquot was injected onto an HPLC with u.v. or fluorescent detection or onto an LC-MS/MS system (see below). To capture potential polar metabolites which may be removed during the extraction process with organic solvents, the supernatant was also directly injected onto these detection systems without further extraction. Negative control incubations were run in parallel that included exclusion of anastrozole, a NADPH-generating system or microsomes (bovine serum albumin was used instead) from the incubation mixture.
LC/MS/MS assay development and metabolite(s) identification
Pilot experiments that were performed using HPLC methods with UV and fluorescent detection to identify any potential anastrozole oxidative metabolites or to determine the disappearance of the parent drug from the microsomal incubates consisting of HLMs and cofactors were noninformative. Although a chromatographic peak that was consistent with an anastrozole oxidative metabolite was identified using the UV detection method, the amount of metabolite formed was too low to perform further kinetic characterization or to conduct structural elucidation. No metabolite peak was detected in the fluorescent scan and anastrozole depletion when incubated with HLMs and cofactors was not substantially different from those observed in negative control incubates. Therefore, a sensitive LC/MS/MS analytical method was developed to identify and quantify human metabolism of anastrozole. Briefly, anastrozole and the internal standard, N-desmethyldiazepam were injected onto an LC-MS/MS that consisted of a LC-20AB pump and SIL-20A HT autosampler (Shimadzu Addison, IL) and an API 2000 LC/MS/MS triple quadruple system (Applied Biosystems Foster City, CA) with an electrospray ion source. Anastrozole and the internal standard were separated on a 100 × 2 mm Luna 3 µ C18(2) 100A stainless steel column (Phenomenex, Torrance, CA). The components were eluted with 50% of 0.1% formic acid in water and 50% of acetonitrile which was degassed in a sonicator for 15 min and delivered at a flow rate of 0.15 ml min−1. The LC eluate was introduced into the ESI source at the same flow rate.
The mass spectrometer was operated using electrospray ionization (ESI) with an ion-spray voltage of +5200 V and temperature was set at 450°C. The positive ion multiple reaction monitoring (MRM) mode analysis was performed using nitrogen as the collision gas. The nitrogen nebulizer gas and curtain gas were both set at 20 psi. The curtain gas (nitrogen) flow and the ion-spray flow were set at 0.6 and 0.9 l min−1, respectively. The pressure in the collision cell was set at 2.20 mTorr. The orifice voltage and ring voltages were set to +35 and +400 V, respectively. A dwell time of 400 ms and a pause time of 5 ms between scans were used to monitor the precursor/product ion pairs.
Identification of metabolites
Microsomal incubate (extracted or unextracted) was injected onto the LC-MS/MS described above. The following approaches were used to identify possible metabolites in microsomal incubates of anastrozole. Because we had no access to anastrozole metabolites (except triazole, which was commercially available), the fragmentation of anastrozole was used as a guide for interpretation and identification of potential anastrozole metabolites, where possible. Targeted precursor ion scans were performed as a first step of metabolite identification. As a guide, a list of expected metabolites was compiled and possible metabolic pathways reconstructed based on the available limited metabolism data in the product label of anastrozole as well as predicted metabolic alterations. Suspected masses identified through the precursor ion scan were targeted for product ion analysis to confirm further the identity of these analytes as true metabolites derived from anastrozole. Based on the data obtained from LC/MS/MS analysis, one metabolite peak that was consistent with hydroxyanastrozole was identified. Two positive transitions (quantifier MRM and confirmed with the qualifier MRM transition) were used for the measurement of anastrozole (+294/225 and +294/115), hydroxy-anastrozole (+310.2/241.5 and +310.2/214.5) and the internal standard desmethyldiazepam (+271/140 and +271/208).
Hydroxyanastrozole was quantified using the ratio of area under the curve (AUC) of the metabolite to AUC of the internal standard, Concentrations of the metabolite were measured by standard curves obtained with anastrozole, as authentic samples of the synthetic metabolite were not available to us. The limitation of this approach is that the MS/MS properties of the metabolite and the parent compound may differ as a result of altered molecular mass. Therefore, the formation rates (pmol min−1 mg−1 protein or pmol min−1 pmol−1 CYP) calculated in this study should be viewed more appropriately as arbitrary units. The limit of detection of the assay was 50 pg ml−1 and the limit of quantification was 100 pg ml−1.
Kinetic analyses in HLMs
Kinetic studies for anastrozole metabolism were conducted at conditions linear for duration of incubation and protein concentration in four different HLM preparations. Anastrozole (1–200 µM) was incubated in duplicate for 30 min at 37°C with HLMs (0.5 mg protein ml−1) and a NADPH-generating system. The reaction was terminated and processed as described above. Apparent kinetic parameters were calculated (see Data analysis).
Correlation analysis of anastrozole hydroxylation
Correlation between apparent formation rate of hydroxyanastrozole and individual P450 activity was tested using a panel of characterized HLMs. Low (1 µm) and high (20 µm) concentrations of anastrozole were incubated with microsomes from different characterized human livers. The activity of each P450 isoform in each HLM preparation was as supplied (CellzDirect) and was determined by isoform-specific reaction markers (http://cellzdirect.com/). The correlation coefficients between the formation rates of anastrozole metabolites and the activity of each CYP isoform in different HLMs were calculated (see Data analysis).
Inhibition studies
Enzyme specific inhibitors were used to obtain further information on the specific P450s involved. Anastrozole (10 µm) was incubated for 30 min at 37°C with HLMs (0.5 mg ml−1) and cofactor in the absence (control) and presence of the following known isoform-specific inhibitors: furafylline (20 µm) for CYP1A2, pilocarpine (50 µm) for CYP2A6, thioTEPA (50 µm) for CYP2B6, quercetin (20 µm) and trimethoprim (50 and 100 µm) for CYP2C8, sulfaphenazole (25 µm) for CYP2C9, ticlopidine (5 µm) for 2C19 and CYP2B6, quinidine (1 µm) for CYP2D6, diethyldithiocarbamate (50 µm) for CYP2E1, troleandomycin (50 µM) and ketoconazole (1 µm) for CYP3A. Inhibition by furafylline, thioTEPA, diethyldithiocarbamate, and troleandomycin has been shown to be time-dependent. Therefore, a preincubation protocol was implemented in which the inhibitor was first incubated with human liver samples and cofactors for 15 min at 37°C before the reaction was initiated by addition of the respective selective substrate probe. The specific conditions used with these inhibitors have been described in detail in our recent publications [12–14]. Percent inhibition of metabolite formation rate by isoform-specific inhibitors was calculated by comparing the inhibited activity with uninhibited controls (without inhibitors).
Anastrozole metabolism by expressed CYP enzymes
The ability of different expressed human CYP enzymes (CYP1A2, 2A6, 2B6, 2C8, 2C9, 2C19, 2D6, 2E1, 3A4, or 3A5) to catalyze anastrozole hydroxylation was tested by incubating anastrozole (20 µm) with 13 pmol P450 and a NADPH-generating system (same composition as above) at 37°C for 30 min. All other incubation conditions and assay of the metabolites were the same as described for HLMs above. Preliminary analysis suggested that CYP3A4 and CYP3A5 showed the highest activity towards anastrozole hydroxylation. Therefore, the kinetics for hydroxyanastrozole formation were performed by incubating increasing concentrations of anastrozole (1–200 µm) at 37°C for 30 min with cofactors and 13 pmol expressed CYP3A4 and CYP3A5 with or without b5. Kinetic parameters were obtained using appropriate enzyme kinetic equations (see Data analysis).
B. Anastrozole metabolism by glucuronidation
The glucuronidation of anastrozole was measured in pooled HLMs, a panel of recombinant UGTs (UGT1A1, 1A3, 1A4, 1A6, 1A7, 1A8, 1A9, 1A10, 2B4, 2B7, 2B15, and 2B17), and in microsomes from 53 individual human livers. Microsomes were pre-incubated with alamethicin (25 µg ml−1 in final incubation) on ice for 15 min. Alamethicin was dissolved in 90/10 incubation buffer/ethanol. The final concentration of ethanol in the reaction was less than 0.1%. Incubations (200 µl) contained anastrozole (500 µm), 50 mm Tris-HCl buffer (pH 7.4), 5 mm MgCl2, 0.5 mg ml−1 microsomes/recombinant UGTs and 5 mm UDP-glucuronic acid (UDPGA), and were carried out at 37°C for 90 min. The reactions were terminated by addition of 0.2 ml acetonitrile containing 10 µm testosterone (internal standard), followed by centrifugation at 20 500 g for 15 min. After drying down supernatant under nitrogen gas at 37°C, samples were reconstituted in mobile phase. Aliquots (40 µl) were then analyzed by LC-MS/MS. Control incubations without UDPGA, substrate or microsomes were carried out to ensure that produced metabolites were microsome- and UDPGA-dependent. For identification of the glucuronide peaks, enzymatic hydrolysis was also carried out. In these experiments, an aliquot of the incubation mixture was mixed with an equal volume of 0.15 m acetate buffer pH 5.0 containing 2000 units of β-d-glucuronidase and incubated for 24 h at 37°C prior to analysis. LC/MS/MS was used for monitoring anastrozole metabolite. The mass spectrometer was an API2000 triple quadrupole (Applied Biosystems, Waltham, MA, USA) equipped with an ESI interface. The cone voltage was 40 V and the capillary temperature was 550°C. Nitrogen was used as nebulizing and auxiliary gas. The nebulizing gas backpressure was set at 40 psi, and auxiliary gas at 20 (arbitrary units). Initially, the mass spectrometer was programmed to perform full scans between m/z 100 and 1000 in order to observe the [M-H]+ signals. Subsequently, the ions monitored for anastrozole glucuronide (analyte) and testosterone (IS) were m/z 470.2/225.2 and m/z 289.2/97.2 in positive ion mode, respectively. The product ion spectra were acquired simultaneously using collision energy of 30 eV with a cone voltage of 40 V. Due to the absence of authentic standards for anastrozole glucuronide, quantification of the glucuronide was accomplished using a standard curve for anastrozole. The standard curve for quantifying anastrozole glucuronidation was linear from 34 to 831 nm, and the correlation coefficient was >0.99. The precision was assessed by relative standard deviation (RSD = SD/Mean × 100%), while the accuracy was calculated as relative mean error of calculated concentrations from nominal concentrations [RME = (calculated concentration − nominal concentration)/nominal concentration × 100%]. The accuracy and precision of the back-calculated values for each concentration were less than 15% of the nominal values. The retention times of anastrozole glucuronide, anastrozole and testosterone were 5.8 min, 6.8 min, and 8.7 min, respectively.
Kinetic studies for anastrozole glucuronidation were carried out in pooled HLMs and recombinant UGT1A4. Anastrozole (10–1000 µm) was preincubated with pooled HLMs/recombinant UGT1A4 (0.5 mg protein ml−1) at 37°C for 5 min in a final volume of 200 µl of 50 mm Tris-HCl buffer (pH 7.5) containing 5 mm MgCl2 and 25 µg ml−1 alamethicin. After preincubation of the reaction mixture, the reaction was started by adding UDPGA for a final concentration of 5 mm. After incubation at 37°C for 90 min, the reaction was terminated by addition of 200 µl acetonitrile containing internal standard (testosterone, 200 nm), followed by centrifugation at 20 500 g for 15 min to obtain the supernatant. Aliquots (40 µl) were then analyzed by LC/MS/MS.
Inhibition studies
Anastrozole (500 µm) was incubated in the absence or presence of different concentrations of hecogenin, a known UGT1A4 inhibitor. The concentration of anastrozole was close to its apparent Km. Incubation was performed for 90 min using 0.5 mg protein ml−1 recombinant UGT1A4 or pooled HLMs.
Correlation analysis of anastrozole glucuronidation
Imipramine N-glucuronidation (UGT1A4) assay:
Imipramine N-glucuronidation activity was determined as previously reported [15] using 0.5 mm imipramine.
Morphine 3-glucuronidation (UGT2B7) assay
Morphine glucuronidation had been previously measured in the same set of liver microsomes [16] using 0.5 mm morphine.
Genotyping
Genotyping of UGT1A4 and UGT2B7 polymorphisms was performed in the entire sample set. Two functional UGT1A4 SNPs (70C>A (P24T) and 142T>G (L48V)), as well as two tag UGT1A4 SNPs (448T>C (150L) and 471C>T (157C)), were genotyped. The forward and reverse primers were located outside the open reading frame to obtain specificity of the amplification reaction. Sequencing primers used were 5′-GGCTGGCCACAGGACTGCTTCTC-3′ (forward) and 5′-GGACACTGTGTGCCCTTAAAGTC-3′ (reverse). PCR products were amplified in a volume of 40 µl containing 10× PCR buffer, 15 mm magnesium sulphate, 2 mm dNTP mix, 10 µm primers, 10 ng µl−1 genomic DNA and 5 U µl−1 Taq DNA polymerase (Promega, Madison, WI). Amplification was performed with an initial denaturing step at 95°C for 10 min followed by seven touchdown cycles of primer annealing and extending. Each cycle included incubating for 20 s at each degree from 65–59°C and 72°C for 50 s, then 72°C for 1 min, as well as 35 cycles amplification of 95°C for 20 s, 58°C for 20 s, and 72°C for 50 s and a final extension step at 72°C for 10 min. PCR products were purified by treatment with shrimp alkaline phosphatase and exonuclease I for 1 h at 37°C, followed by 15 min at 72°C to inactivate enzymes. Haplotype-tagging UGT2B7 polymorphisms were also genotyped. The same set of human liver samples had been previously genotyped for UGT2B7 at position 802C>T (H268Y) and the haplotype 4 polymorphisms [16].
Western blot
UGT1A3 and UGT1A4 expression levels (mg protein) in the recombinant microsomes were measured by Western blot using a pan-UGT1A antibody. A UGT2B7-specific antibody was used for measuring protein in UGT2B7 recombinant microsomes. Protein expression (mg) was calculated based on a standard curve constructed using purified UGT1A1 protein as previously published [17].
C. Anastrozole metabolism in vivo
Plasma samples obtained from postmenopausal women with breast cancer who were taking 1 mg day−1 of the drug were used to study anastrozole metabolism in vivo. Women with breast cancer who met eligibility criteria were prescribed 1 mg day−1 anastrozole to be taken orally. Anastrozole was provided by Astra Zeneca. Plasma samples were obtained at 1, 6, and 12 months after starting anastrozole intake. Patients were instructed not to take their dose of anastrozole for that day until after the blood was drawn. Since anastrozole has a mean terminal elimination half-life of ∼50 h in postmenopausal women, it is unlikely that time of sampling post dose will have a significant effect on concentrations. The study was approved by the Institutional Review Board of the Johns Hopkins School of Medicine and written informed consent was provided by each patient before entry to the study.
The LC-MS/MS method described above for microsomal studies was slightly modified to measure anastrozole and hydroxyanastrozole in human plasma [9]. In brief, to 250 µl of plasma samples, the internal standard desmethyldiazepam was added and extracted using ethyl acetate under alkaline pH (0.5 ml of 0.5 m NaOH/glycine buffer, adjusted to pH 10). The sample was vortex mixed and centrifuged for 15 min at 2500 rev min−1 in a Beckman GS-6R centrifuge. The organic layer was then removed and evaporated to dryness. The residue was reconstituted with 50 µl of 0.1% formic acid in water from which 25 µl was injected onto the LC-MS/MS system (see above). To assess the potential role of glucuronidation in the metabolism of anastrozole and/or its metabolite, plasma samples were deconjugated using β-glucuronidase. Plasma samples (250 µl) were incubated for 18 h with 20 µl of 1000 units ml−1β-glucuronidase, 200 µl of 200 mm acetate buffer and 10 µl of 600 mm sodium azide. After the addition of the internal standard the samples were extracted and processed as above. The separation system and the conditions of the LC-MS/MS assay were the same as those described for microsomal studies.
Plasma concentrations of anastrozole were quantified using the ratio of area under the curve (AUC) of anastrozole to AUC of the internal standard and calibration curves that were constructed by spiking blank plasma with known amounts of anastrozole. The limit of detection was 50 pg ml−1 and the limit of quantification was 100 pg ml−1. The concentrations of the metabolite presented in this paper should be viewed as apparent concentrations (arbitrary units ml−1 plasma).
Data analysis
Apparent kinetic parameters (Km and Vmax) for anastrozole hydroxylation and glucuronidation were estimated by fitting apparent formation rates of metabolite vs. anastrozole concentrations to a single- or two-site Michaelis-Menten enzyme kinetic equation using nonlinear regression analysis software (GraphPad Prism Software Inc, Version 5, San Diego, CA) or the Enzyme Kinetics Module of SigmaPlot (Systat Software, Inc., San Jose, CA, USA). Correlation analyses were performed using a Spearman's rank or a Pearson's correlation test with GraphPad Prism Software Version 5 (GraphPad Software Inc., San Diego, CA). P < 0.05 was regarded as statistically significant and all tests were two-sided. Data are presented as mean ± SD or as the mean of duplicate measurements, unless stated otherwise.
Results
Anastrozole oxidation
Identification of anastrozole metabolite in HLM incubates
Using HPLC with UV detection, a single chromatographic peak that was unique to incubations consisting of anastrozole, HLMs and cofactors was noted. This metabolite peak depended on the NADPH-generating system, duration of incubation and microsomal protein and substrate concentrations (data not shown). LC/MS/MS analysis was performed to characterize further this metabolite peak. Precursor ion scanning (ESI conditions) identified that this metabolite exhibited 309 amu (with [M-H]+ at m/z 310) in the anastrozole microsomal incubate that consisted of HLMs and cofactors (but not in negative control experiments) in addition to the 293 amu (with [M-H]+ at m/z 294) of anastrozole (data not shown). To gain further structural information, a MS/MS spectrum of this potential metabolite was obtained by collusion-induced dissociation of the molecular [M+H]+ ion at 310 m/z. The spectrum contained parent ion (m/z 310) and two major characteristic fragment ions, m/z 241.5 and 214.5. These transitions ions were selected in the MRM mode for quantification of the metabolite where the quantifier was measured with the quantifier MRM (m/z 310/241.5) and confirmed with the qualifier MRM transition (m/z 310/214.5). A representative ion chromatogram of the anastrozole metabolite is shown in Figure 2. Together, these data suggested that this metabolite peak was consistent with the monohydroxylated metabolite of anastrozole. In the product label of the drug, hydroxyanastrozole glucuronide has been stated to be one of anastrozole metabolites in human [8]. Although theoretically anastrozole can undergo hydroxylation in the aromatic ring, it is less likely. We believe that the metabolite identified in this study represents methyl-hydroxylation of anastrozole. However, no synthetic metabolite standard was available to us and thus unambiguous assignment of the precise structure could not be made. The exact position of hydroxylation remains to be identified as the metabolite identification was based entirely on mass spectral analysis, because NMR analysis was not performed in this study.
Figure 2.
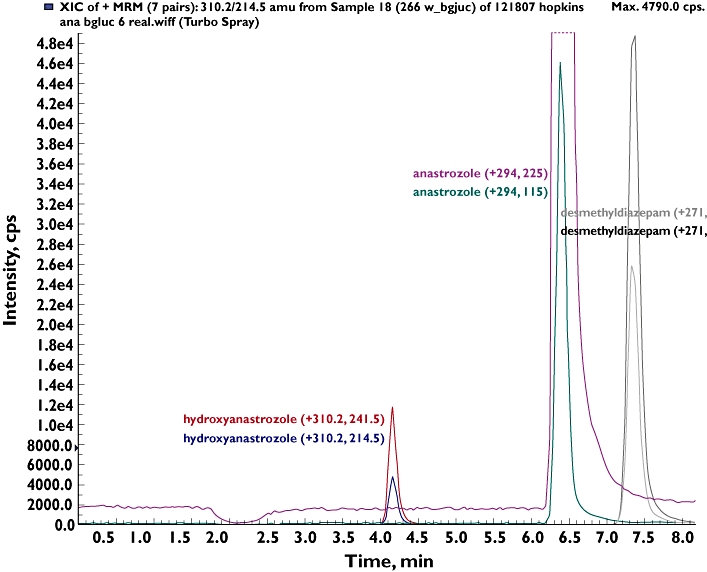
Representative Multiple Reaction Monitoring (MRM) trace chromatograms of anastrozole and its mono-hydroxylated metabolite after incubation of anastrozole (20 µm) in human liver microsomal incubations. Quantifier and qualifier MRMs are shown. The internal stand was desmethyldiazepam
Kinetic analysis in HLMs
Kinetic analysis for the formation of hydroxyanastrozole from anastrozole was studied in four different HLM preparations. A representative kinetic profile of anastrozole metabolism to hydroxyanastrozole in HLMs is shown in Figure 3. The formation rate of hydroxyanastrozole was best fitted to two-site hyperbolic kinetic equations. Accordingly the apparent formation rate of hydroxyanastrozole from anastrozole (0.5–200 µm) in HLMs exhibited biphasic kinetic behaviour in Eadie-Hofstee plots (Figure 3), which suggested the involvement of at least two enzymatic activities that were best described by high-affinity (Km1 and Vmax1) and low-affinity (Km2 and Vmax2) components. The estimated kinetic parameters for the high affinity component are demonstrated in Table 1. The intrinsic metabolic clearance of the high-affinity component (Vmax1/Km1) was 5–10-fold higher than that of the low-affinity component (Vmax2/Km2), and Km2 was much higher (millimolar range) than Km1 (data not shown). Since the metabolite standard was not available to us, quantification of this metabolite was made using standard curves generated with anastrozole. Thus the Vmax values provided in Table 1 and those described in the subsequent tests are not actual values but instead they should be viewed as relative values (arbitrary pmol product min−1 mg−1 protein or arbitrary pmol product min−1 pmol−1 P450).
Figure 3.
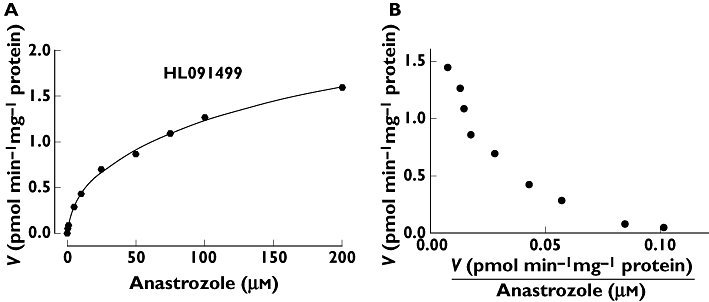
Representative enzyme kinetic plots for the formation of hydroxyanastrozole in three human liver microsomes (HL091499). Anastrozole (0–200 µm) was incubated for 30 min at 37°C with HLMs (0.5 mg ml−1). Each microsomal incubation experiment was carried out in duplicate
Table 1.
Enzyme kinetic parameters of anastrozole metabolite formation in three human liver samples
| Enzyme kinetic parameters* | |||
|---|---|---|---|
| HLMs | Km± SE (µm) | Vmax± SE (pmol min−1 mg−1 protein) | Vmax/Km (µl min−1 mg−1 protein) |
| HL091499 | 4.5 ± 0.43 | 0.39 ± 0.24 | 0.086 |
| SD117 | 20.7 ± 0.15 | 0.65 ± 0.096 | 0.031 |
| SD004 | 21.70 ± 0.92 | 0.63 ± 0.10 | 0.029 |
| HLM (pooled) | 23.5 ± 0.77 | 0.30 ± 0.03 | 0.013 |
| Mean (±SD) | 17.6 ± 8.8 | 0.49 ± 0.17 | 0.040 ± 0.032 |
Computer generated SE is provided for the individual HLMs. Other goodness of fits (r2) in the four HLMs were all greater than 0.98.
Correlation analysis
Apparent rates of anastrozole (1 µm and 20 µm) hydroxylation in a panel of 11 pre-phenotyped HLMs were characterized by interindividual variability (Figure 4). The average apparent hydroxyanastrozole formation rate at 1 µm and 20 µm of anastrozole was 0.098 ± 0.09 pmol min−1 mg−1 protein (range 0.0048–0.258, 53.8 fold) and 0.86 ± 0.55 (range 0.05–1.68, 33-fold). Formation rates of hydroxyanastrozole from 1 and 20 µm anastrozole correlated significantly with the activity of CYP3A, as measured by both testosterone 6β-hydroxylation (Spearman r = 0.93 and 0.96, P = 0.0027 and P = 0.0005, respectively) and midazolam 1′-hydroxylation (Spearman r = 0.86 and 0.89, P = 0.014 and P = 0.007, respectively). Formation rates of anastrozole from 1 µm anastrozole also significantly correlated with CYP2C8 activity (Spearman r = 0.89, P = 0.019), but this did not reach a statistically significant level at the higher substrate concentration (Spearman r = 0.77, P = 0.07) (Table 2).
Figure 4.
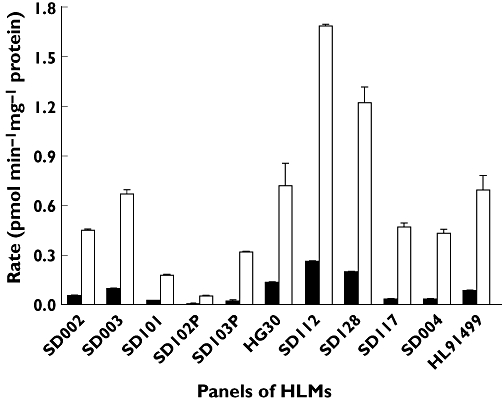
Formation rate of hydroxyanastrozole in a panel of characterized HLMs. Anastrozole (1 µm or 20 µm) was incubated for 30 min at 37°C with HLMs (0.5 mg ml−1). Each microsomal incubation experiment was carried out in duplicate. 1 µm ( ); 20 µm (□)
); 20 µm (□)
Table 2.
Correlation between formation rate of hydroxyanastrozole and individual P450-isoform contents in a panel of HLMs
| Correlation analysis (1 µm anastrozole) | Correlation analysis (20 µm anastrozole) | |||
|---|---|---|---|---|
| CYPs | Spearman r | P value | Spearman r | P value |
| CYP1A2 | 0.05406 | 0.9084 | 0.01802 | 0.9694 |
| CYP2A6 | 0.07143 | 0.879 | 0.1071 | 0.8192 |
| CYP2B6 | 0.3214 | 0.4821 | 0.25 | 0.5887 |
| CYP2C8 | 0.8857 | 0.0188 | 0.7714 | 0.0724 |
| CYP2C9 | 0.2857 | 0.5345 | 0.3929 | 0.3833 |
| CYP2C19 | 0.1786 | 0.7017 | 0.03571 | 0.9394 |
| CYP2D6 | 0.4325 | 0.3325 | 0.5225 | 0.2289 |
| CYP2E1 | −0.2143 | 0.6445 | −0.3571 | 0.4316 |
| CYP3A (T) | 0.9266 | 0.0027 | 0.9636 | 0.0005 |
| CYP3A (M) | 0.8571 | 0.0137 | 0.8929 | 0.0068 |
T, testosterone 6β-hydroxylation; M, midazolam 1′-hydroxylation.
Chemical inhibition of anastrozole oxidation
To identify the P450 enzymes participating in anastrozole hydroxylation, 10 µm anastrozole was incubated with P450 isoform-specific inhibitors with HLMs and cofactors. As shown in Figure 5, troleandomycin (50 µm) and ketoconazole (1 µm) showed strong inhibitory effect on rates of hydroxyanastrozole formation (by 80% and 95%, respectively), while thioTEPA (50 µm), quercetin (20 µm) and pilocarpine (50 µm) were moderate inhibitors (by 46%, 54% and 60%, respectively). Since quercetin showed some inhibition and since the correlation analysis implicated CYP2C8 in anastrozole metabolism, trimethoprim, a selective inhibitor of CYP2C8 [14] was used as additional inhibitor probe. Trimethoprim (50 µm) inhibited anastrozole metabolism by only 11.3% (Figure 5) and at 100 µm by only 22% (data not shown). A similar inhibition profile was observed when 1 µm anastrozole was used (by 26% and 33% at 50 and 100 µm trimethoprim). The inhibitory effect of other isoform-specific inhibitors was marginal (by <20%).
Figure 5.
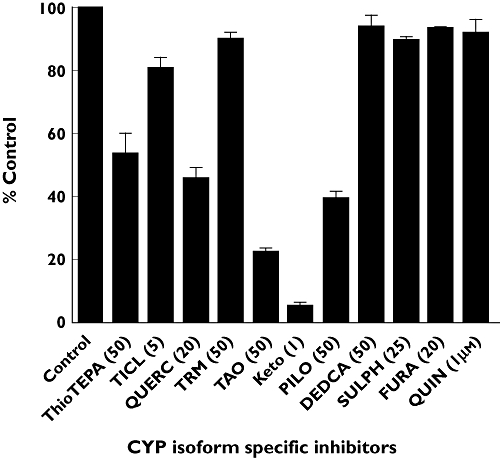
Inhibition of hydroxyanastrozole formation from anastrozole by a panel of CYP enzyme specific inhibitors. The final concentrations of the inhibitors used are indicated in parentheses. Anastrozole (10 µm) was incubated for 30 min at 37°C with HLMs (0.5 mg ml−1) in the presence or absence of chemical inhibitors. Each microsomal incubation experiment was carried out in duplicate. Abbreviations: TICL ticlopidine (CYP2B6 and CYP2C19), QUERC quercetin (CYP2C8), TMP trimethoprim (CYP2C8), TAO troleandomycin (CYP3A), Keto ketoconazole (CYP3A), PILO pilocarpine (CYP2A6), DEDCA diethyldiothicarbamate (CYP2E1), SULPH sulfaphenazole (CYP2C9), FURA furafylline (CYP1A2), QUIN quinidine
Metabolism of anastrozole by expressed CYPs
Because there was a strong correlation between formation rate of hydroxyanastrozole from 1 µm anastrozole and from 20 µm anastrozole (Spearman r = 0.96, P = 0.0005), anastrozole (20 µm) was used in the experiments involving expressed CYPs (Figure 6). CYP3A4 and CYP3A5 catalyzed anastrozole (20 µm) hydroxylation at the highest rate (0.0023 pmol min−1 pmol−1 P450 and 0.0026 pmol min−1 pmol−1 P450, respectively) (Figure 6A). CYP2B6, CYP2D6 and CYP2C8 also formed hydroxyanastrozole, but when compared with CYP3A4 and 3A5, the involvement of these enzymes was very low (10.8-, 25- and 30-fold lower, respectively) (Figure 6A). Full kinetic analyses for hydroxyanastrozole formation were carried out for CYP3A4, CYP3A5 and CYP2C8 (Figure 6B). The apparent hydroxyanastrozole formation rates fit better to a single-site Michaelis-Menten equation. The kinetic parameters are shown in Table 3. In addition, the kinetics of anastrozole hydroxylation by CYP3A4 and CYP3A5 was performed in the presence of b5. The estimated Km and Vmax values, respectively, for these two enzymes were 13.9 µm and 0.02 pmol pmol−1 P450 × min for CYP3A4 and 21.5 µm and 0.01 pmol min−1 pmol−1 P450 for CYP3A5 in the presence of cytochrome b5. As shown in Table 3, the apparent formation rate of hydroxyanastrozole by CYP3A4 and CYP3A5 was 170- and 4.6-fold higher in those preparations with b5 than those without b5 (the Km value was increased by 3.7 and decreased by 2.3-fold, respectively). The Km and Vmax values for formation of hydroxyanastrozole by expressed CYP2C8 was 86.8 µm and 0.00005 pmol min−1 pmol−1 P450, respectively.
Figure 6.
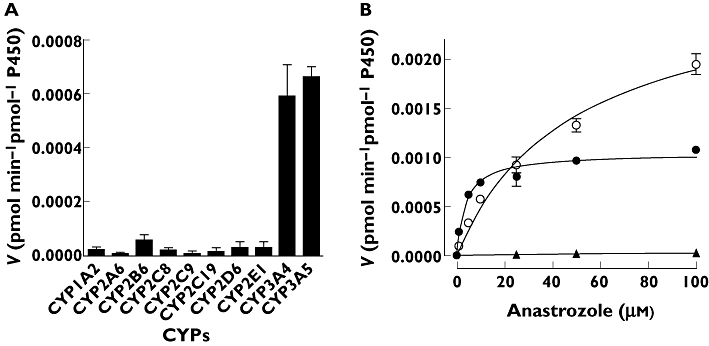
Metabolism of anastrozole to hydroxyanastrozole by expressed human CYP enzymes. (A) Formation rate of hydroxyanastrozole from anastrozole in a panel of expressed human CYPs. Anastrozole (20 µm) was incubated for 30 min at 37°C with each isoform (52 pmol ml−1). Each microsomal incubation experiment was carried out in duplicate. (B) Representative enzyme kinetic plots for hydroxyanastrozole formation by CYP3A4, CYP3A5 and CYP2C8 in the absence of cytochrome b5 (b). Anastrozole (0–100 µm) was incubated for 30 min at 37°C with each isoform (52 pmol ml−1). Each microsomal incubation experiment was carried out in duplicate. (B) CYP3A4 ( ); CYP3A5 (
); CYP3A5 ( ); CYP2C8 (
); CYP2C8 ( )
)
Table 3.
Enzyme kinetic parameters of anastrozole metabolite formation in CYP3A4 and CYP3A5 with and without cytochrome b5
| Enzyme kinetic parameters | |||
|---|---|---|---|
| Expressed enzymes | Km (µm) | Vmax (pmol min−1 pmol−1 P450 | Vmax/Km (µl min−1 pmol−1 P450) |
| CYP2C8 | 86.8 | 0.00005 | <0.00001 |
| CYP3A4 (−b5) | 3.7 | 0.001 | 0.00027 |
| CYP3A5 (−b5) | 48.5 | 0.003 | 0.00006 |
| CYP3A4 (+b5) | 13.9 | 0.017 | 0.0012 |
| CYP3A5 (+b5) | 21.5 | 0.013 | 0.0006 |
Anastrozole glucuronidation
Anastrozole glucuronidation in HLMs
Anastrozole has been observed to form a quaternary ammonium linked triazole N+-glucuronide metabolite [18]. To characterize the role of UGT enzymes in anastrozole glucuronidation, the glucuronidation of anastrozole was measured in pooled HLMs. The highest rate of anastrozole glucuronide formation after normalization by protein content was observed with UGT1A4 (mean anastrozole glucuronide formation = 3863 nmol min −1mg−1), followed by UGT1A3 (150 pmol min−1 mg−1), and UGT2B7 (57 pmol min−1 mg−1) (Figure 7A).
Figure 7.
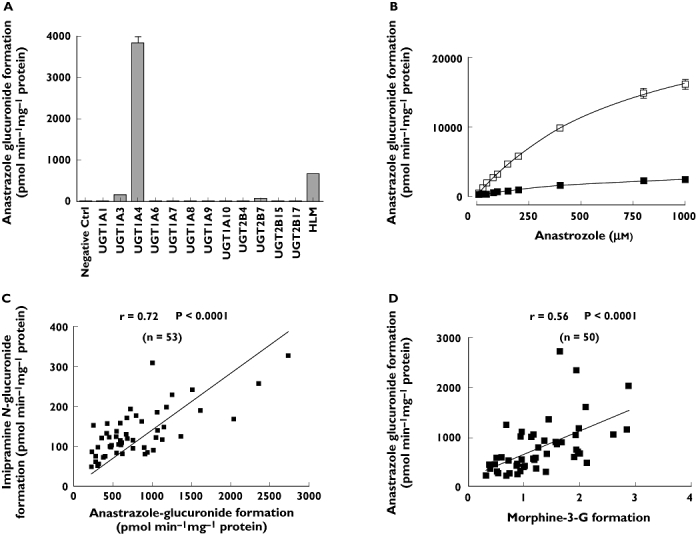
Anastrozole glucuronidation in HLMs and UGTs enzymes. (A) Formation rate of anastrozole glucuronide from anastrozole single concentration in a panel of purified human UGTs. Incubations were performed with 0.5 mg ml−1 microsomal protein and 500 µm anastrozole for 90 min at 37°C. Data represent means of duplicate determinations from a single experiment. (B) Kinetics of anastrozole glucuronidation by recombinant human UGT1A4 and pooled HLMs. Anastrozole (10–1000 µm) was incubated with pooled HLMs and UGT1A4 at 37°C for 90 min. Data represent the mean of duplicate determinations. (C) Pearson correlation between anastrozole glucuronide formation rate and imipramine glucuronidation (UGT1A4 probe substrate) formation rate in a bank of human liver samples (n = 53). (D) Pearson correlation between anastrozole glucuronide formation rate and morphine 3-glucuronidation (UGT2B7 probe substrate) formation rate in a bank of human liver samples (n = 50). (B) recombinant UGT1A4 (□); pooled HLM ( )
)
Kinetic analysis of anastrozole glucuronidation by pooled HLMs and expressed human UGT1A4
Kinetic studies were performed using pooled HLMs and expressed human UGT1A4. The apparent kinetic parameters (Km and Vmax) in HLMs were estimated to be 540.4 ± 69.1 µm and 3.7 ± 0.2 nmol min−1 mg−1, respectively, comparable with parameters determined in a previous report [10]. As anastrozole glucuronidation seems to be specifically catalyzed by UGT1A4, kinetic studies were performed in expressed UGT1A4. The Km and Vmax values in expressed human UGT1A4 were 768.6 ± 73.9 µm and 28.8 ± 1.5 pmol min−1 mg−1, respectively (Figure 7B). The Km value for anastrozole glucuronidation derived from HLMs was close to that obtained in expressed UGT1A4.
Inhibition of anastrozole glucuronidation by UGT1A4 inhibitor
The inhibition of anastrozole glucuronidation by hecogenin, a known UGT1A4 inhibitor, was observed in both pooled HLMs and recombinant UGT1A4. Hecogenin at 5 µm inhibited anastrozole glucuronidation activity by 38.4% in pooled HLMs (P < 0.01) and 62.2% in expressed UGT1A4 (P < 0.01). At 50 µm, hecogenin reduced anastrozole glucuronide formation by 69.3% in pooled HLMs (P < 0.01) and 83.6% in expressed UGT1A4 (P < 0.01).
Correlation study
As shown in Figure 7C, significant correlations were observed between anastrozole glucuronide formation and UGT1A4 activity, as measured by imipramine- N-glucuronidation (Pearson r = 0.72, P < 0.0001). Significant correlations were also observed between anastrozole glucuronide formation with morphine 3-glucuronidation (Pearson r = 0.56, P < 0.0001), a probe reaction for UGT2B7 (Figure 7D). Morphine 3-glucuronidation formation activities were not available for 3 of 53 samples.
UGT1A4 is polymorphically expressed in human livers [19]. UGT1A4 P24T (UGT1A4*2) and L48V (UGT1A4*3) genetic variants have been shown to be associated with reduced glucuronidation activities and have minor allele frequencies of 8% and 9%, respectively [20]. To test whether these polymorphisms (along with others) accounted for some of the wide interindividual variability observed in our panel of human liver microsomes, we carried out an exploratory genotype-phenotype association analysis between the most common genetic variants in UGT1A4 and UGT2B7 and anastrozole glucuronidation formation rates. Three out of 53 samples were excluded from genotyping analysis due to insufficient amounts of DNA. We found no statistically significant association between these genetic variants and anastrozole glucuronide formation rates in the 50 human liver microsomes (supporting Table S1). However, there are limitations to these data. First, they were unlikely to have adequate power to detect genotype-phenotype associations given the small sample size studied and multiple SNPs with small functional effects were tested. Second, we realize that UGT1A4 activity may be compromised by the alternative splicing variant previously discovered [21], which may confound the association between the polymorphisms and UGT1A4 activity. Future studies in large sample sizes and a complete set of SNPs might be warranted.
Identification of anastrozole metabolites in vivo
To test the relevance of the in vitro findings to in vivo anastrozole metabolism, anastrozole and the metabolites were measured in plasma samples obtained from women with breast cancer (n = 54) who were taking anastrozole (1 mg day−1) for 6 or 12 months. To evaluate the extent of conjugation by UGTs in vivo, anastrozole and hydroxyanastrozole were measured with and without deconjugation using β-glucuronidase. In β-glucuronidase untreated samples, the mean (±SD) plasma concentrations of anastrozole and hydroxyanastrozole were 47.5 ± 21.8 ng ml−1 (range 10.1–119.7, 11-fold) and 0.056 ± 0.029 ng ml−1 (range 0.0024–0.037, 15-fold), respectively. In β-glucuronidase treated patient samples, anastrozole and hydroxyanastrozole concentrations in plasma were 47.1 ± 17.1 ng ml−1 (range 17.0–90.9, 5-fold) and 0.62 ± 0.27 ng ml−1 (range 0.22–1.5, 7-fold), respectively. Plasma concentrations of anastrozole in β-glucuronidase treated samples were not statistically different than those untreated with β-glucuronidase (Figure 8A; P = 0.82). However, the average plasma concentration of hydroxyanastrozole was significantly higher (>11-fold) in the β-glucuronidase samples compared with those samples untreated with β-glucuronidase (Figure 8B; P < 0.0001).
Figure 8.
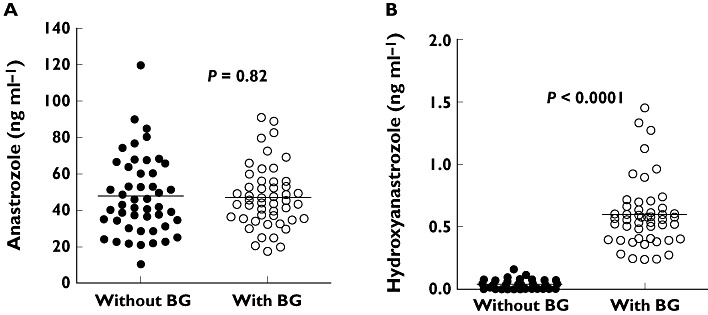
Plasma concentrations of anastrozole (A) and hydroxyanastrozole (B) in postmenopausal women with breast cancer taking 1 mg day−1 anastrozole orally. Six or 12 month plasma samples were analyzed without β-glucuronidase (BG) and after incubation with β-glucuronidase (BG). (A) Without BG (•); With BG (○); (B) Without BG (•); With BG (○)
Discussion
In this study, we report the first systematic characterization of anastrozole oxidation and glucuronidation in vitro and in vivo. We show that anastrozole is oxidized to hydroxyanastrozole predominantly by CYP3A4. Once formed, this metabolite undergoes extensive glucuronidation in vivo. We also show that anastrozole is directly glucuronidated in vitro predominantly by UGT1A4, although the contribution of this route in vivo remains to be clarified through measurement of metabolites in urine. These data serve as a basis to design clinical drug interaction and pharmacogenetic studies with CYP3A4 and UGT1A4 aimed at predicting anastrozole clearance.
Available information suggests that anastrozole is predominantly cleared by hepatic metabolism via oxidation and glucuronidation in humans to pharmacologically inactive metabolites that include triazole, hydroxyanastrozole (free and glucuronide conjugated) as well as anastrozole N-glucuronide [8–10]. In the present study, we have identified anastrozole N-glucuronide, hydroxyanastrozole and conjugated hydroxyanastrozole using in vitro and in vivo approaches. In our in vitro experiments, we identified anastrozole glucuronide, supporting the notion that anastrozole can undergo direct glucuronidation (probably N-glucuronidation) [8–10]. In addition, we have shown that anastrozole is hydroxlated to form hydroxyanastrozole. The precise structure of the hydroxylated and glucuronidated metabolites remains unknown. The structural identification relied solely on MS/MS data and no NMR analysis was performed. Hydroxylation seems to occur at the methyl site (http://med.astrazeneca.co.jp/product/IF/ARI_IF.pdf), but the possibility of hydroxylation at the aromatic or methylene sites cannot be ruled out [10]. The N-glucuronidation could take place at one of the nitrogen atoms in the triazole ring [10]. Triazole (a potential product of N-dealkylation of anastrozole) has been considered to be the major circulating metabolite in plasma of patients taking anastrozole [8]. However, this or any other N-dealkylated metabolites of anastrozole [e.g. 3,5-Bis-(2-methylpropiononitrile)-benzoic acid] could not be detected in our in vitro studies. The reason for this discrepancy is not clear. We had access to synthetic triazole standard from commercial sources, but repeated attempts to develop an assay for this metabolite did not materialize, which could be due to its low molecular weight and high polarity. The experimental conditions that led to the conclusion that triazole is the main metabolite in plasma have not been published [8] and thus are difficult to evaluate. Given the robust and sensitive LC-MS/MS assay and the different approaches we used to study anastrozole metabolism, it seems unlikely that this is due to limitations of our experimental or assay conditions. Together, it appears that anastrozole hydroxylation represents the main oxidative metabolic pathway and anastrozole N-glucuronidation represents the main conjugation pathway of the drug. The role anastrozole N-dealkylation plays in anastrozole metabolism could not be confirmed under the experimental conditions used in the present study.
To test that the identified oxidative and glucuronidated metabolic routes of anastrozole in vitro are also present in vivo, the plasma concentrations of anastrozole and its metabolites were measured in postmenopausal women with early-stage breast cancer who were taking anastrozole (1 mg day−1). Plasma samples were assayed without and with β-glucuronidase treatment to reflect free (unconjugated) and total (unconjugated + conjugated) concentrations, respectively. Consistent with our in vitro data, hydroxyanastrozole was detected and quantified in plasma mainly as the conjugated and, to a small extent, as the unconjugated (free) form, suggesting that once formed by oxidation, the hydroxylated metabolite is then subsequently efficiently conjugated probably by UGTs. This metabolite represented the main circulating metabolite of anastrozole, with wide interpatient variability. As with our in vitro findings, no traces of triazole or any other metabolites of anastrozole (free or conjugated) were found in any of the clinical samples assayed. Our in vivo data suggest that exposure of anastrozole glucuronide (or more appropriately anastrozole conjugate) in plasma is negligible from the present data and from a previous report [9]], as shown by the lack of significant difference between the free and total anastrozole. However, these data do not exclude the possibility that direct anastrozole conjugation (N-glucuronidation) is an important pathway for anastrozole metabolism in vivo. First, it is possible that enzyme hydrolysis using β-glucuronidase may be incomplete for N-glucuronide metabolites [22]. Second, determinations of anastrozole urinary metabolites that will provide insight in to the contribution of this pathway to anastrozole elimination were not performed in this study.
Our data strongly suggest that CYP3A4 is the dominant enzyme catalyzing anastrozole hydroxylation. The data supporting this conclusion include significant correlation of CYP3A activity with anastrozole hydroxylation in a panel of HLMs, CYP3A-isoform specific inhibitors reduce anastrozole hydroxylation by over 80%, expressed CYP3A4 catalyzed anastrozole hydroxylation at the highest rate and the Km values obtained from HLMs were close to those values obtained from expressed CYP3A4. Of note, CYP3A has been shown to hydroxylate triazole derivative compounds such as alprazolam (4-hydroxylation), voriconazole (methyl-hydroxylation), and estazolam (4-hydroxylation) [20, 23, 24]. Although involvement of CYP3A5 is suggested from the expressed kinetic data, the Km value was much higher than that of CYP3A4. Also, correlation analyses and inhibition with quercetin (a nonspecific inhibitor of CYP2C8) implicated CYP2C8 in anastrozole hydroxylation, but data from expressed CYP2C8 show marginal activity towards this reaction, with Km values over 85 µm, and the extent of inhibition by trimethoprim, a more selective inhibitor, was minimal. Together, the contribution of CYP2C8 is likely to be negligible, if any.
Using enzyme kinetic and correlation studies along with expressed UGTs, we have demonstrated that anastrozole glucuronidation is predominantly catalyzed in vitro by UGT1A4 (and to a lesser extent by UGT2B7 and UGT1A3). These findings are consistent with those of Lazarus & Sun [10] who, citing their unpublished results, reported that UGT1A4 was the main enzyme responsible for anastrozole N-glucuronidation in vitro, with an average Km value of ∼637 µm, which was close to the results of the present study.
Anastrozole is currently considered as an established cost-effective agent for first-line endocrine treatment of all stages of hormone receptor positive breast cancer in postmenopausal women and is being tested as a chemopreventive agent in postmenopausal women at risk for developing the disease [5, 8], but its pharmacodynamic activity varies widely among individuals [9]. This is evidenced by the fact that, some patients develop serious side effects that include musculoskeltal symptoms [e.g. arthralgia (∼35% of patients)] and fractures (∼10% of patients), etc.], whereas others do not [5]. Also not all patients show beneficial response to anastrozole therapy. The reasons for such wide interpatient variability in response to anastrozole are likely to be multiple. The present data derived from spot plasma concentrations suggest that anastrozole exposure among postmenopausal patients may vary widely and this, in turn, may contribute to differences in the effects of anastrozole. Thus, as a first step to allow testing of the role drug exposure plays in variable drug response, we identified the specific pathways and the specific enzymes involved in anastrozole metabolism. Our in vitro data together with in vivo evidence suggest that anastrozole hydroxylation to hydroxyanastrozole by CYP3A4, which then undergo rapid and extensive conjugation probably by UGTs, and anastrozole N-glucuronidation to anastrozole glucuronide predominantly by UGT1A4 are the principal metabolic pathways of anastrozole (Figure 9). It is well known that CYP3A4 is expressed in the liver and in extra-hepatic tissues (e.g. gut, breast), with extensive interindividual variability mostly dictated by environmental factors, age, gender and concomitant drug administration [25]. Studies on drug interactions and pharmacogenetic determinants on anastrozole exposure are generally limited. Tamoxifen appears to reduce anastrozole exposure by ∼27% [26]. The precise mechanisms involved are not known, but it is possible that this interaction is mediated through induction of CYP3A4 by tamoxifen. In vitro studies suggest that tamoxifen activates nuclear receptors such as the human pregnane X receptor (PXR) and induces CYP3A4 in primary human hepatocytes [27, 28]. Although CYP3A5 is polymorphically expressed among different ethnic groups, with CYP3A5*3 (inactive gene form) allele frequencies of 30%, 50% and 90% in African, Asian and Caucasian, respectively [29, 30], the role of CYP3A5 and any SNPs in the gene that codes for this enzyme in anastrozole metabolism in vivo is likely to be small because the in vitro kinetic data presented here suggest CYP3A4 (not CYP3A5) is the dominant enzyme responsible for anastrazole metabolism at therapeutically relevant concentrations. We have also shown (at least in vitro) that UGT1A4 is the main enzyme responsible direct glucuronidation of anastrozole, although the precise contribution of this enzyme to the in vivo metabolism of the drug is not yet fully known. Of note, the UGT1A4 gene is highly polymorphic [20, 21, 31, 32]. Future studies related to the impact of CYP3A4 drug–drug interaction and/or UGT1A4 genetic polymorphisms on anastrozole drug disposition among postmenopausal women with breast cancer would be of clinical value.
Figure 9.
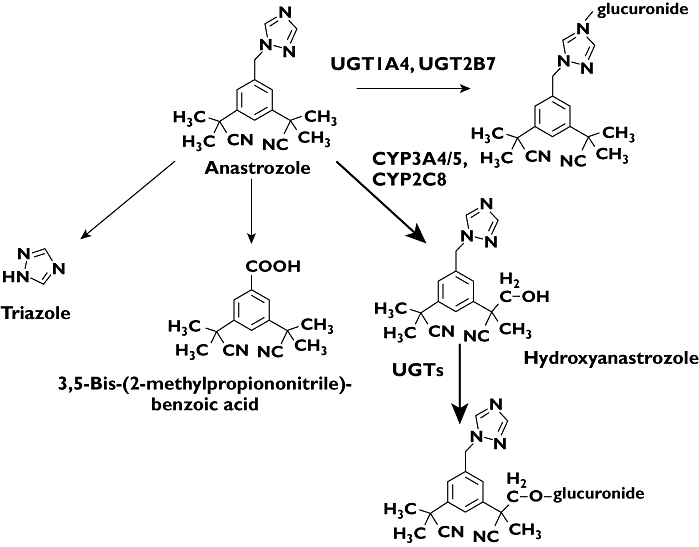
In vitro biotransformation pathways of anastrozole to its primary and secondary metabolites and the specific CYP and UGT enzymes involved. The principal enzymes responsible for each eliminating pathway are indicated. The relative contribution of each pathway to the overall metabolism of anastrozole is shown by the thickness of the arrow, and the principal CYP enzymes responsible are highlighted in bold
Acknowledgments
This work was supported by Award Numbers 5R01CA118981 from the National Cancer Institute, Award Number 5R01GM078501-04 from the National Institute of General Medical Sciences, Award Numbers 5U01GM061373 [Consortium for Breast Cancer Pharmacogenomics (COBRA)] and U01 GM61393 [the Pharmacogenetics of Anticancer Agents Research (PAAR) Group] from the National Institute Of General Medical Sciences, Bethesda, MD, and by the Breast Cancer Research Foundation (Vered Stearns). Data will be deposited into PharmGKB (http://pharmgkb.org/) (supported by Award Number U01GM61374 from the National Institute of General Medical Sciences). AstraZeneca provided anastrozole pills for the purpose of the clinical study. We also thank Dr Moshe Finel from the Center for Drug Research, Faculty of Pharmacy, University of Helsinki, Helsinki, Finland, for performing the experiments related with UGT1A10.
Competing interests
VS and DAF received investigator-initiated grants from Pfizer and Novartis. VS received remuneration from Astra Zeneca for giving a talk. The terms of this arrangement are being managed by the Johns Hopkins University in accordance with its conflict of interest policies. DAF is a member of the Scientific Advisory Board of Medco Health Solutions Inc., a pharmaceuticals benefits management company.
Supporting Information
Additional Supporting information may be found in the online version of this article:
Table S1 Effect of candidate UGT SNPs on the formation rate of anastrozole glucuronide in a panel of 50 human livers.
Please note: Wiley-Blackwell are not responsible for the content or functionality of any supporting materials supplied by the authors. Any queries (other than missing material) should be directed to the corresponding author for the article.
REFERENCES
- 1.Parkin DM, Bray F, Ferlay J, Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55:74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 2.American Cancer Society. Breast cancer facts and figures 2009–2010. 2010:1–36. [Google Scholar]
- 3.Osborne CK. Tamoxifen in the treatment of breast cancer. N Engl J Med. 1998;339:1609–18. doi: 10.1056/NEJM199811263392207. [DOI] [PubMed] [Google Scholar]
- 4.Smith IE, Dowsett M. Aromatase inhibitors in breast cancer. N Engl J Med. 2003;348:2431–42. doi: 10.1056/NEJMra023246. [DOI] [PubMed] [Google Scholar]
- 5.Forbes JF, Cuzick J, Buzdar A, Howell A, Tobias JS, Baum M. Effect of anastrozole and tamoxifen as adjuvant treatment for early-stage breast cancer: 100-month analysis of the ATAC trial. Lancet Oncol. 2008;9:45–53. doi: 10.1016/S1470-2045(07)70385-6. [DOI] [PubMed] [Google Scholar]
- 6.Coombes RC, Hall E, Gibson LJ, Paridaens R, Jassem J, Delozier T, Jones SE, Alvarez I, Bertelli G, Ortmann O, Coates AS, Bajetta E, Dodwell D, Coleman RE, Fallowfield LJ, Mickiewicz E, Andersen J, Lønning PE, Cocconi G, Stewart A, Stuart N, Snowdon CF, Carpentieri M, Massimini G, Bliss JM, the Intergroup Exemestane Study A randomized trial of exemestane after two to three years of tamoxifen therapy in postmenopausal women with primary breast cancer. N Engl J Med. 2004;350:1081–92. doi: 10.1056/NEJMoa040331. [DOI] [PubMed] [Google Scholar]
- 7.The Breast International Group (BIG) 1-98 Collaborative Group. A comparison of letrozole and tamoxifen in postmenopausal women with early breast cancer. N Engl J Med. 2005;353:2747–57. doi: 10.1056/NEJMoa052258. [DOI] [PubMed] [Google Scholar]
- 8.Sanford M, Plosker GL. Anastrozole: a review of its use in postmenopausal women with early-stage breast cancer. Drugs. 2008;68:1319–40. doi: 10.2165/00003495-200868090-00007. [DOI] [PubMed] [Google Scholar]
- 9.Ingle JN, Buzdar AU, Schaid DJ, Goetz MP, Batzler A, Robson ME, Northfelt DW, Olson JE, Perez EA, Desta Z, Weintraub RA, Williard CV, Flockhart DA, Weinshilboum RM. Variation in anastrozole metabolism and pharmacodynamics in women with early breast cancer. Cancer Res. 2010;70:3278–86. doi: 10.1158/0008-5472.CAN-09-3024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lazarus P, Sun D. Potential role of UGT pharmacogenetics in cancer treatment and prevention: focus on tamoxifen and aromatase inhibitors. Drug Metab Rev. 2010;42:176–88. doi: 10.3109/03602530903208652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bradford MM. A rapid and sensitive method for the quantitation of microgram quantities of protein utilizing the principle of protein-dye binding. Anal Biochem. 1976;72:248–54. doi: 10.1016/0003-2697(76)90527-3. [DOI] [PubMed] [Google Scholar]
- 12.Ogburn ET, Jones DR, Masters AR, Xu C, Guo Y, Desta Z. Efavirenz primary and secondary metabolism in vitro and in vivo: identification of novel metabolic pathways and cytochrome P450 (CYP) 2A6 as the principal catalyst of efavirenz 7-hydroxylation. Drug Metab Dispos. 2010;38:1218–29. doi: 10.1124/dmd.109.031393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Jeong S, Nguyen PD, Desta Z. Comprehensive in vitro analysis of voriconazole inhibition of eight cytochrome P450 (CYP) enzymes: major effect on CYPs 2B6, 2C9, 2C19, and 3A. Antimicrob Agents Chemother. 2009;53:541–51. doi: 10.1128/AAC.01123-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wen X, Wang JS, Backman JT, Laitila J, Neuvonen PJ. Trimethoprim and sulfamethoxazole are selective inhibitors of CYP2C8 and CYP2C9, respectively. Drug Metab Dispos. 2002;30:631–5. doi: 10.1124/dmd.30.6.631. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y, Ramirez J, House L, Ratain MJ. Comparison of the drug-drug interactions potential of erlotinib and gefitinib via inhibition of UDP-glucuronosyltransferases. Drug Metab Dispos. 2010;38:32–9. doi: 10.1124/dmd.109.029660. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Innocenti F, Liu W, Fackenthal D, Ramírez J, Chen P, Ye X, Wu X, Zhang W, Mirkov S, Das S, Cook E, Jr, Ratain MJ. Single nucleotide polymorphism discovery and functional assessment of variation in the UDP-glucuronosyltransferase 2B7 gene. Pharmacogenet Genomics. 2008;18:683–97. doi: 10.1097/FPC.0b013e3283037fe4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Yoder Graber AL, Ramirez J, Innocenti F, Ratain MJ. UGT1A1*28 genotype affects the in-vitro glucuronidation of thyroxine in human livers. Pharmacogenet Genomics. 2007;17:619–27. doi: 10.1097/FPC.0b013e3280121fe9. [DOI] [PubMed] [Google Scholar]
- 18.McCann DJ, Dyroff MC Hall JE. Isolation, identification and synthesis of a novel quaternary ammonium linked triazole glucuronide metabolite of the aromatase inhibitor anastrozole. 1997. In: 8th North American ISSX Meeting, Hilton Head, SC, October 1997: ISSX Proceedings, 1997, Hilton Head, SC, 1997: abstract 224.
- 19.Wiener D, Fang JL, Dossett N, Lazarus P. Correlation between UDP-glucuronosyltransferase genotypes and 4-(methylnitrosamino)-1-(3-pyridyl)-1-butanone glucuronidation phenotype in human liver microsomes. Cancer Res. 2004;64:1190–6. doi: 10.1158/0008-5472.can-03-3219. [DOI] [PubMed] [Google Scholar]
- 20.Ehmer U, Vogel A, Schutte JK, Krone B, Manns MP, Strassburg CP. Variation of hepatic glucuronidation: novel functional polymorphisms of the UDP-glucuronosyltransferase UGT1A4. Hepatology. 2004;39:970–7. doi: 10.1002/hep.20131. [DOI] [PubMed] [Google Scholar]
- 21.Girard H, Levesque E, Bellemare J, Journault K, Caillier B, Guillemette C. Genetic diversity at the UGT1 locus is amplified by a novel 3′ alternative splicing mechanism leading to nine additional UGT1A proteins that act as regulators of glucuronidation activity. Pharmacogenet Genomics. 2007;17:1077–89. doi: 10.1097/FPC.0b013e3282f1f118. [DOI] [PubMed] [Google Scholar]
- 22.Kassahun K, Mattiuz E, Franklin R, Gillespie T. Olanzapine 10-N-glucuronide. A tertiary N-glucuronide unique to humans. Drug Metab Dispos. 1998;26:848–55. [PubMed] [Google Scholar]
- 23.Murayama N, Imai N, Nakane T, Shimizu M, Yamazaki H. Roles of CYP3A4 and CYP2C19 in methyl hydroxylated and N-oxidized metabolite formation from voriconazole, a new anti-fungal agent, in human liver microsomes. Biochem Pharmacol. 2007;73:2020–6. doi: 10.1016/j.bcp.2007.03.012. [DOI] [PubMed] [Google Scholar]
- 24.Pai HV, Upadhya SC, Chinta SJ, Hegde SN, Ravindranath V. Differential metabolism of alprazolam by liver and brain cytochrome (P4503A) to pharmacologically active metabolite. Pharmacogenomics J. 2002;2:243–58. doi: 10.1038/sj.tpj.6500115. [DOI] [PubMed] [Google Scholar]
- 25.Wilkinson GR. Drug metabolism and variability among patients in drug response. N Engl J Med. 2005;352:2211–21. doi: 10.1056/NEJMra032424. [DOI] [PubMed] [Google Scholar]
- 26.Dowsett M, Cuzick J, Howell A, Jackson I. Pharmacokinetics of anastrozole and tamoxifen alone, and in combination, during adjuvant endocrine therapy for early breast cancer in postmenopausal women: a sub-protocol of the ‘Arimidex and tamoxifen alone or in combination’ (ATAC) trial. Br J Cancer. 2001;85:317–24. doi: 10.1054/bjoc.2001.1925. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Sane RS, Buckley DJ, Buckley AR, Nallani SC, Desai PB. Role of human pregnane X receptor in tamoxifen- and 4-hydroxytamoxifen-mediated CYP3A4 induction in primary human hepatocytes and LS174T cells. Drug Metab Dispos. 2008;36:946–54. doi: 10.1124/dmd.107.018598. [DOI] [PubMed] [Google Scholar]
- 28.Desai PB, Nallani SC, Sane RS, Moore LB, Goodwin BJ, Buckley DJ, Buckley AR. Induction of cytochrome P450 3A4 in primary human hepatocytes and activation of the human pregnane X receptor by tamoxifen and 4-hydroxytamoxifen. Drug Metab Dispos. 2002;30:608–12. doi: 10.1124/dmd.30.5.608. [DOI] [PubMed] [Google Scholar]
- 29.Kuehl P, Zhang J, Lin Y, Lamba J, Assem M, Schuetz J, Watkins PB, Daly A, Wrighton SA, Hall SD, Maurel P, Relling M, Brimer C, Yasuda K, Venkataramanan R, Strom S, Thummel K, Boguski MS, Schuetz E. Sequence diversity in CYP3A promoters and characterization of the genetic basis of polymorphic CYP3A5 expression. Nat Genet. 2001;27:383–91. doi: 10.1038/86882. [DOI] [PubMed] [Google Scholar]
- 30.Wojnowski L, Kamdem LK. Clinical implications of CYP3A polymorphisms. Expert Opin Drug Metab Toxicol. 2006;2:171–82. doi: 10.1517/17425255.2.2.171. [DOI] [PubMed] [Google Scholar]
- 31.Saeki M, Saito Y, Jinno H, Sai K, Ozawa S, Kurose K, Kaniwa N, Komamura K, Kotake T, Morishita H, Kamakura S, Kitakaze M, Tomoike H, Shirao K, Tamura T, Yamamoto N, Kunitoh H, Hamaguchi T, Yoshida T, Kubota K, Ohtsu A, Muto M, Minami H, Saijo N, Kamatani N, Sawada JI. Haplotype structures of the UGT1A gene complex in a Japanese population. Pharmacogenomics J. 2006;6:63–75. doi: 10.1038/sj.tpj.6500335. [DOI] [PubMed] [Google Scholar]
- 32.Saeki M, Saito Y, Jinno H, Sai K, Hachisuka A, Kaniwa N, Ozawa S, Kawamoto M, Kamatani N, Shirao K, Minami H, Ohtsu A, Yoshida T, Saijo N, Komamura K, Kotake T, Morishita H, Kamakura S, Kitakaze M, Tomoike H, Sawada J. Genetic variations and haplotypes of UGT1A4 in a Japanese population. Drug Metab Pharmacokinet. 2005;20:144–51. doi: 10.2133/dmpk.20.144. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


