Abstract
Glycosylphosphatidylinositol (GPI) anchoring provides an alternative to transmembrane domains for anchoring proteins to the cell surface in eukaryotes. GPI anchors are synthesized in the endoplasmic reticulum via the sequential addition of monosaccharides, fatty acids, and phosphoethanolamines to phosphatidylinositol. Deficiencies in GPI biosynthesis lead to embryonic lethality in animals and to conditional lethality in eukaryotic microbes by blocking cell growth, cell division, or morphogenesis. We report the genetic and phenotypic analysis of insertional mutations disrupting SETH1 and SETH2, which encode Arabidopsis homologs of two conserved proteins involved in the first step of the GPI biosynthetic pathway. seth1 and seth2 mutations specifically block male transmission and pollen function. This results from reduced pollen germination and tube growth, which are associated with abnormal callose deposition. This finding suggests an essential role for GPI anchor biosynthesis in pollen tube wall deposition or metabolism. Using transcriptomic and proteomic approaches, we identified 47 genes that encode potential GPI-anchored proteins that are expressed in pollen and demonstrated that at least 11 of these proteins are associated with pollen membranes by GPI anchoring. Many of the identified candidate proteins are homologous with proteins involved in cell wall synthesis and remodeling or intercellular signaling and adhesion, and they likely play important roles in the establishment and maintenance of polarized pollen tube growth.
INTRODUCTION
Glycosylphosphatidylinositol (GPI) membrane anchors provide an alternative to transmembrane domains for anchoring proteins to the cell surface in eukaryotes. GPI anchoring can confer localized or polarized targeting and therefore can dramatically alter the functional properties of proteins. In animals, GPI-anchored proteins (GAPs) include a broad range of cell surface proteins, such as enzymes, receptors, complement regulators, and adhesion molecules (Ikezawa, 2002). In plants, GAPs form a relatively abundant class of proteins present in the plasma membrane (Sherrier et al., 1999). Many of the demonstrated and predicted Arabidopsis GAPs show similarity to characterized cell surface proteins as diverse as β-1,3-glucanases, metalloproteases and aspartylproteases, glycerophosphodiesterases, phytocyanins, multi-copper oxidases, extensins, classic arabinogalactan proteins, plasma membrane receptors, peptides, and lipid transfer–like proteins (Borner et al., 2002, 2003).
To date, only 4 of the 248 predicted Arabidopsis GAPs (Borner et al., 2002, 2003) have been functionally investigated. Mutations in the COBRA gene (Roudier et al., 2002) affect the orientation of cell expansion in the root (Schindelman et al., 2001), whereas the multi-copper oxidase–related protein SKU5, shown to possess a GPI anchor, is involved in the control of directional root growth (Sedbrook et al., 2002). Mutations in the pectate lyase–like protein PMR6 alter cell wall structure and pectin content and confer mildew susceptibility (Vogel et al., 2002). Mutations of the SOS5 locus confer salt hypersensitivity to roots, resulting in abnormal cell expansion and growth arrest under salt stress (Shi et al., 2003). The phenotypes of these mutants suggest that GAPs might be especially important in aspects of cell wall remodeling and directional cell expansion.
GPI anchors are synthesized in the endoplasmic reticulum via the sequential addition of monosaccharides, fatty acids, and phosphoethanolamines to phosphatidylinositol (reviewed by Schultz et al., 2000; Ikezawa, 2002). The proteins that catalyze this pathway have been well studied in mammalian cells, yeast, and protozoa, but essentially nothing is known about them in plants. The first step involves the transfer of N-acetylglucosamine to phosphatidylinositol (PI) by GPI-N-acetylglucosaminyltransferase (GPI-GnT), an enzymatic complex comprising at least six subunits (PIG-A, PIG-C, PIG-H, GPI-1, PIG-P, and DPM2). The product formed is deacetylated subsequently by PIG-L to yield glucosamine-PI. Three mannose residues, donated by dolichol-phosphate-mannose (DPM), are added sequentially to the anchor. The addition of phosphorylethanolamine on the third mannose residue is catalyzed by PIG-O and PIG-F. The removal of the hydrophobic C-terminal sequence and the attachment of the GPI anchor to the ω site of candidate proteins involves the GAA1 and GPI8 proteins. In the Golgi apparatus, the GPI core structure in animals and yeast is elaborated further by the addition of sugar side chains on mannose residues and often by remodeling of the lipid to a ceramide. Although GPI anchor biosynthesis has not been studied in plants, Oxley and Bacic (1999) determined that the GPI anchor of a Pyrus communis arabinogalactan protein has a minimal core oligosaccharide structure analogous to those found in animals, protozoa, and yeast. However, it is distinguished by the presence of a β(1-4)-galactosyl substitution of the 6-linked mannose residue.
GPI biosynthetic pathways have been studied extensively in animals and microbes and are a target for the development of parasite-specific therapeutic agents. Deficiencies in GPI biosynthesis lead to embryonic lethality in animals and to conditional lethality in eukaryotic microbes by blocking cell growth, cell division, or morphogenesis. Although mammalian cells can survive in culture without GPI anchors, partial deficiency at different steps of the GPI biosynthetic pathway can have drastic effects in whole animals or in eukaryotic microbes. In human, an X-linked mutation of PIG-A in hematopoietic stem cells leads to paroxysmal nocturnal hemoglobinuria, an acquired clonal disease (Rosti, 2000). In haploid Saccharomyces cerevisiae strains, PIG-C mutations affect vegetative growth such that ascospores carrying the mutation germinate but complete no more than four cell divisions (Leidich et al., 1995). Partial deficiency of DPM biosynthesis in human causes the congenital disorder of glycosylation, resulting in seizures, hypotonia, and dysmorphic features (Imbach et al., 2000). Moreover, side chain addition mutations have drastic effects on GPI protein transport, remodeling, and cell wall integrity in yeast (Benachour et al., 1999; Gaynor et al., 1999) but only partially affect the expression of GPI-anchored surface receptors in murine embryonal carcinoma cells (Hong et al., 1999). However, the effects of such mutations on embryogenesis or cell differentiation are unknown.
Here, we report the genetic and phenotypic analysis of insertional mutations disrupting the SETH1 and SETH2 genes that encode homologs of PIG-C and PIG-A, respectively, two components of the GPI-GnT complex. Heterozygous GPI biosynthetic knockout mutations in Arabidopsis have no effect on sporophytic development and megagametogenesis but show male gametophyte–specific effects that almost completely block transmission through pollen. GPI-deficient pollen grains develop normally and are viable at pollination but show reduced germination and pollen tube growth. To provide a genome-wide view of GAPs expressed in pollen, we used both transcriptomic and proteomic approaches. We identified 47 genes whose transcripts were expressed in mature pollen and that encode probable GAPs. Eleven of these proteins were confirmed to be associated with pollen membranes through GPI anchoring. Our data suggest that pollen-expressed GAPs fulfill essential functions in pollen germination and tube growth.
RESULTS
seth1 Insertional Mutations Specifically Block Male Gametophytic Transmission
From a genetic screen of 3616 DsE and DsG transposon lines based on marker segregation ratio distortion, we isolated 19 independent gametophytic mutations (E. Lalanne, C. Michaelidis, A. Johnson, R. Patel, R. Howden, J. Moore, W. Gagliano, J.P. Vielle Calzada, U. Grossniklaus, and D. Twell, unpublished data), including an insertion, seth1-1, that disrupts a putative phosphatidylinositol-glycan synthase subunit C gene. Two additional insertional alleles, seth1-2 and seth1-3, were identified in the Syngenta collection (Sessions et al., 2002). Heterozygous seth1 insertions had no effect on sporophytic development, and no homozygous mutant plants were identified by segregation analysis of progeny from >500 plants derived from seth1-1 heterozygotes. Analysis of self-progeny revealed that all three seth1 alleles segregated 1:1 for the marker within the insertion, suggesting reduced transmission of the gametes harboring the insertion (Table 1). Progeny testing revealed strict cosegregation of antibiotic or herbicide resistance with the reduced genetic transmission phenotype for all three seth1 alleles (data not shown). Genetic transmission of seth1 mutations through the male and female gametes was determined in reciprocal test crosses of heterozygous mutants and wild-type plants. The transmission efficiency of the mutant allele describes the fraction of mutant alleles that are transmitted successfully to the progeny (Howden et al., 1998). Female transmission was normal, but there was almost no transmission (transmission efficiency < 1%) of seth1 alleles through the male, demonstrating that all three seth1 mutations specifically affect male gametophyte development and/or functions (Table 1).
Table 1.
Genetic Transmission Analysis of seth1 and seth2 Mutations
| Mutant | Selfing
|
Mutant as Female Parent |
Mutant as Male Parent |
|||
|---|---|---|---|---|---|---|
| KanR:KanS | Ratio | KanR:KanS | TE Female | KanR:KanS | TE Male | |
| seth1-1 | 2235:2098 | 1.07 | 151:162 | 93.2% | 0:412 | 0% |
| seth1-2 | 289:285 | 1.01 | 185:139 | 133% | 3:592 | 0.67% |
| seth1-3 | 231:225 | 1.03 | 306:289 | 105.8% | 4:396 | 1% |
| seth2 | 91:85 | 1.07 | 140:148 | 94.5% | 8:503 | 1.6% |
Numbers of progeny producing wild-type and mutant pollen are shown together with the calculated transmission efficiency (TE = number of mutant progeny/number of wild-type progeny × 100) through male and female gametes. Self and reciprocal crosses between heterozygous mutants and the wild type are depicted. KanR, kanamycin-resistant; KanS, kanamycin-sensitive.
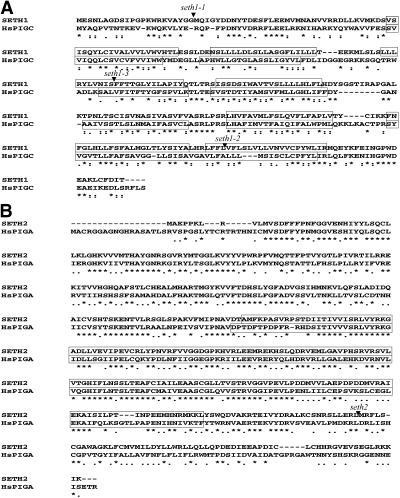
In the three seth1 insertion lines, the DsG transposon (seth1-1) or T-DNAs (seth1-2 and seth1-3) was inserted into the single exon of At2g34980 (SETH1) (Figure 1A). SETH1 encodes a protein that is 31.6% identical (52.7% similar) over 296 amino acids to human PIG-C (HsPIGC). No other related proteins were identified in the Arabidopsis genome. Given that mammalian PIG-C and its known homologs in other organisms are relatively divergent in sequence (21.5% identity and 38.4% similarity between HsPIGC and its yeast homolog GPI2), SETH1 likely encodes the Arabidopsis homolog of PIG-C. PIG-C and GPI2 are associated with the endoplasmic reticulum and show multiple conserved hydrophobic segments, suggesting that they are integral membrane proteins (Inoue et al., 1996). Using topology prediction programs, we identified six to eight potential transmembrane segments in HsPIGC and SETH1, six of which were conserved using the TmHMM_v2 algorithm (http://www.cbs.dtu.dk/services/TMHMM-2.0/).
Figure 1.
Alignment of the Predicted Amino Acid Sequences of Arabidopsis and Human Proteins.
(A) Arabidopsis SETH1 and human PIG-C (HsPIGC).
(B) Arabidopsis SETH2 and human PIG-A (HsPIGA).
Potential transmembrane domains predicted by the TmHMM_v2 algorithm (http://www.cbs.dtu.dk/services/TMHMM-2.0/) are boxed in (A). Arrowheads indicate the positions of the seth1-1, seth1-2, seth1-3, and seth2 insertions. The glycosyltransferase group 1 domain predicted by SMART (http://smart.embl-heidelberg.de/) is boxed in (B). Stars indicate identical amino acid residues, and double dots indicate similar amino acid residues. Deletions/insertions are indicated by dashes. Sequence alignments were made using CLUSTAL W.
Disruption of the GPI-GnT Catalytic Subunit PIG-A Has a Male Gametophyte–Specific Effect
To further investigate the role of GPI anchor biosynthesis in pollen, we identified a T-DNA insertion, seth2, in At3g45100 that encodes a putative homolog of the GPI-GnT catalytic subunit PIG-A (Figure 1B). SETH2 also is a single-copy gene in Arabidopsis and encodes a protein that is 47.8% identical (64.4% similar) over 494 amino acids to its human homolog PIG-A (Miyata et al., 1993). The seth2 mutation was found to result in a male-specific gametophytic defect similar to those of all three seth1 alleles, showing only residual male transmission of 1.6% (Table 1). The insertion in seth2 likely completely blocks GPI-GnT enzyme activity and GPI anchor biosynthesis. The residual transmission of seth1 and seth2 mutant alleles suggests that the gametophytic synthesis of GPI is essential for pollen function but that a few pollen tubes are able to reach and fertilize ovules in the absence of gametophytically synthesized GPIs.
GPI-Deficient Pollen Grains Develop Normally
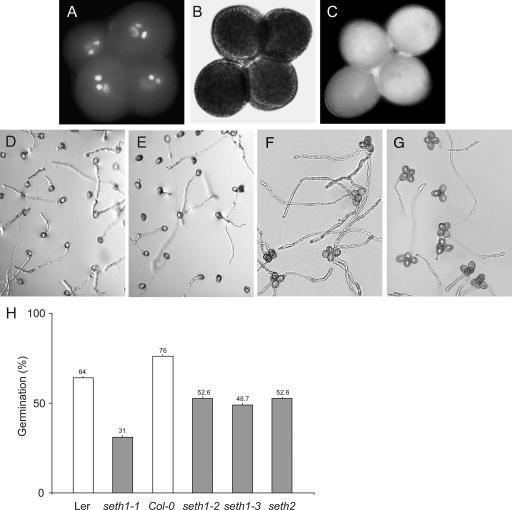
The mature pollen phenotypes of seth1 and seth2 plants were analyzed by 4′,6-diamidino-2-phenylindole staining and fluorescence microscopy. Mutant pollen was similar in size and appearance to wild-type pollen. The seth1-3 mutant was homozygous for the qrt1 mutation (Preuss et al., 1994), and plants with the genotype qrt1/qrt1;+/seth1-3 produced tetrads containing two wild-type and two seth1-3 mutant pollen grains. All pollen grains within tetrads from seth1-3 heterozygotes showed a uniform size and correctly differentiated vegetative and sperm cell nuclei (Figure 2A). Alexander staining (Alexander, 1969) revealed no cytoplasmic or pollen wall abnormalities (Figure 2B). Vital staining using fluorescein diacetate (Figure 2C) showed that >96% of mature pollen grains were viable. Moreover, aniline blue staining did not show the abnormal callose accumulation (data not shown) that has been observed in other male gametophytic mutants, such as gemini pollen1 (Park and Twell, 2001) and raring-to-go (Johnson and McCormick, 2001). No differences in cellulose or pectin staining were observed in mature pollen of heterozygous mutant plants using calcofluor white or ruthenium red staining, respectively. We conclude that GPI biosynthetic mutations have no obvious effects on pollen development or plasma membrane integrity or viability. Because <2% ovule or seed (n = 400) abortion phenotypes were observed in siliques derived from self or male outcrosses, prefertilization effects, including defective pollen tubes occupying the micropyle, or postfertilization effects on seed development cannot account for the reduced male transmission of seth1 and seth2 mutations.
Figure 2.
Pollen Morphology and in Vitro Germination Efficiency for the Wild Type and seth1 and seth2 Heterozygotes.
(A) to (C) Mature tetrads of qrt1/qrt1;+/seth1-3 pollen stained with 4′,6-diamidino-2-phenylindole (A), Alexander stain (B), and fluorescein diacetate (C).
(D) to (G) In vitro germination of pollen from wild-type +/+ (D), +/seth2 (E), qrt1/qrt1;+/+ (F), and qrt1/qrt1; +/seth1-3 (G) plants.
(H) Histogram showing the percentage in vitro germination of pollen from heterozygous seth1-1, seth1-2, seth1-3, and seth2 plants and their respective wild-type controls (n > 1000 pollen grains for each genotype). Standard errors for six independent experiments are shown. Col-0, Columbia; Ler, Landsberg erecta.
seth1 and seth2 Mutations Affect Pollen Germination and Tube Growth
We tested the pollen germination efficiency of seth1 and seth2 mutants using in vitro germination assays (Figures 2D to 2G). Pollen from wild-type control plants showed germination efficiencies that ranged from 64 to 76%. By contrast, only 31 to 52.6% of pollen from heterozygous mutants was capable of germination (Figure 2H), suggesting that mutant pollen is partially defective for germination in vitro.
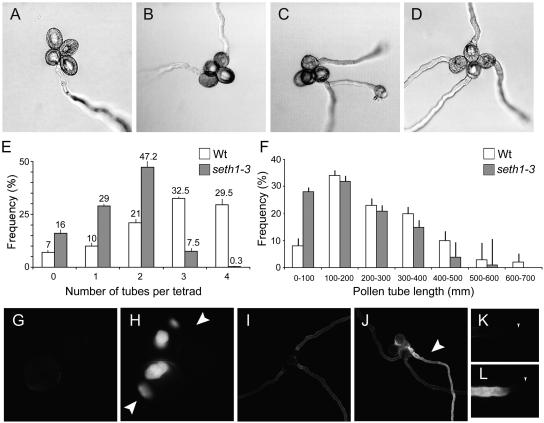
seth1-3 allowed tetrad analysis of pollen germination ability in vitro. Tetrads containing one to four germinated pollen grains were scored (Figures 3A to 3D). In wild-type control plants (qrt1/qrt1;+/+), 32.5% of tetrads produced three tubes and 29.5% of tetrads produced four tubes (Figure 3E), whereas in qrt1/qrt1;+/seth1-3 plants, only 7.5% of tetrads had three tubes and very few tetrads produced four tubes (0.3%). These data confirm the primary outcome of defective gametophytic GPI synthesis to be reduced pollen germination efficiency in vitro.
Figure 3.
In Vitro Germination of Wild-Type qrt1/qrt1;+/+ and Mutant qrt1/qrt1;+/seth1-3 Tetrads.
(A) to (D) qrt1/qrt1;+/+ tetrads showing one to four pollen tube(s).
(E) Percentage of wild-type (qrt1/qrt1;+/+) and mutant (qrt1/qrt1;+/seth1-3) tetrads showing zero to four pollen tube(s) after 16 h. Standard errors for six independent experiments are shown (n > 1000 pollen grains for each genotype). Wt, wild type.
(F) Pollen tube lengths for wild-type (qrt1/qrt1;+/+) and mutant (qrt1/qrt1;+/seth1-3) tetrads showing three pollen tubes (n = 180 pollen grains for each mutant).
(G) and (H) Nongerminated qrt1/qrt1;+/+ (G) and qrt1/qrt1;+/seth1-3 (H) tetrads. Arrows indicate spores showing abnormal callose accumulation.
(I) and (J) qrt1/qrt1;+/+ (I) and qrt1/qrt1;+/seth1-3 (J) tetrads producing three tubes. The arrowhead indicates a tube showing abnormal callose deposition.
(K) and (L) Tips of wild-type (K) and seth1-3 (L) pollen tubes. Tip ends are indicated by arrowheads.
To determine if GPI deficiency has an effect on pollen tube elongation, we measured the lengths of pollen tubes from tetrads producing three pollen tubes, reasoning that at least one tube produced by +/seth1-3 tetrads must carry a mutant seth1-3 allele (Figure 3F). The mean pollen tube length was 258 μm in the wild type and 185 μm in +/seth1-3 tetrads, representing a 28% reduction in mean tube length in +/seth1-3. When the frequency distribution of tube lengths was plotted, 28% of +/seth1-3 tetrads produced pollen tubes of <100 μm, whereas wild-type tetrads showed only 8% of tubes of <100 μm. These data demonstrate a further role for GPI-GnT components in pollen tube elongation in vitro.
To investigate potential cell wall composition defects associated with in vitro germination and growth failures, we stained wild-type and mutant pollen with calcofluor white, ruthenium red, and aniline blue to detect cellulose, pectin, and callose, respectively. No abnormalities were detected in cellulose or pectin staining. By contrast, intense patches of callose staining (Figure 3H) were observed in nongerminated pollen from seth1-3 heterozygotes. In the wild type, only 4.5% of ungerminated pollen grains showed abnormal callose deposition, whereas 13.5% of pollen from seth1-3 heterozygotes showed very strong callose staining (n = 400). Tetrads producing three tubes (where at least one is known to carry a mutant allele) were examined further. In 85% (n = 40) of tetrads from seth1-3 heterozygotes producing three pollen tubes, one tube showed high levels of abnormal callose deposition, with irregular banding patterns transverse to the pollen tube axis (Figure 3J). Mutant pollen tube tips also showed abnormal callose staining (Figure 3L). Similar phenotypes were observed in seth1-2 and seth2 mutants but not in pollen tubes from wild-type tetrads that produced three tubes (n = 60).
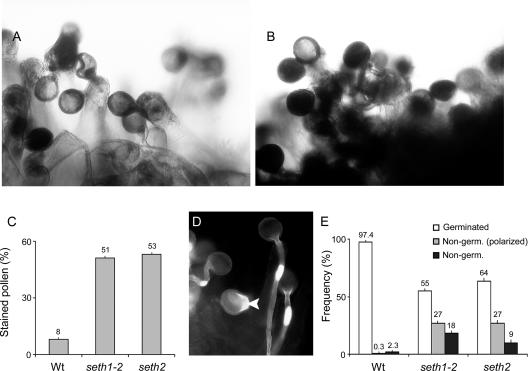
Reduced germination and tube growth efficiency in seth2 were confirmed using in vivo pollination assays (Figure 4). To avoid the potentially complicating effects of stigma maturity and emasculation stresses, pollination was performed on stigmas of male-sterile plants (ms1-1/ms1-1). Pistils of ms1-1 plants were confirmed to respond similarly to those of the wild type or seth2 heterozygotes in competitive pollinations, such that seth2 transmission on ms1-1 pistils was 1.7% (n = 120).
Figure 4.
In Vivo Germination of Wild-Type, seth1-2, and seth2 Pollen Grains.
The morphology and frequency of in vivo germination were monitored using Alexander ([A] to [C]) and aniline blue ([D] and [E]) staining after pollination of ms1-1 stigmas.
(A) and (B) Pollen grains from the wild type (A) and seth2 heterozygotes (B) at 4 h after pollination.
(C) Histogram showing the percentage Alexander staining for pollen of wild type (Wt), +/seth1-2, and +/seth2 plants at 4 h after pollination (n > 1000 pollen grains for each genotype). Standard errors for three independent experiments are shown.
(D) Fluorescence micrograph of an aniline blue–stained stigma showing germinated and polarized (arrowhead) wild-type pollen 2 h after pollination.
(E) Histogram showing the frequency of germinated, polarized, and nonpolarized pollen grains at 2 h after pollination with pollen from wild-type, +/seth1-2, and +/seth2 plants (n = 400 pollen grains for each genotype). Standard errors for three independent experiments are shown.
We developed an in vivo pollination assay to initially measure the overall efficiency of pollen germination and tube growth in vivo. This involved pollinating excised pistils harvested from male-sterile (ms1-1) plants and subsequently treating pistils with Alexander stain (Alexander, 1969). This process allowed pollen germination and tube growth to be scored based on the presence or density of the cytoplasm (Figures 4A and 4B). In the wild type, only 8% of pollen grains attached to the papillar cells were strongly stained at 4 h after pollination, showing that most had transferred their cytoplasm into the pollen tube. By contrast, 51 and 53% of pollen grains from seth1-2 and seth2 heterozygotes, respectively, were strongly stained after the same period (Figure 4C). This resulted from a complete failure to germinate or failure to establish sufficiently long pollen tubes to ensure complete translocation of the cytoplasm out of the pollen grains.
Pollen germination and tube growth also were assayed at 2 h after pollination by staining fixed pistils with aniline blue (Figures 4D and 4E). On ms1-1 stigmas pollinated with wild-type pollen, 97.4% of pollen grains had developed a pollen tube and 1% had initiated germination, indicated by the local outgrowth of the pollen wall and/or the polarized deposition of callose (Figure 4D). By contrast, in pollinations with pollen from seth1-2 heterozygotes, only 55% of pollen grains developed a pollen tube, 27% had initiated germination, and 18% showed no evidence of polarized growth (Figure 4E). A slightly greater germination efficiency (64%) was observed with pollen from seth2 heterozygotes, but 36% failed to germinate (Figure 4E). We conclude that GPI biosynthetic mutations disturb pollen germination and tube growth in planta but that a proportion of GPI-deficient pollen grains are able to support the development of a pollen tube on stigmatic papillae.
Limited Pollination Does Not Restore Male Transmission in GPI-Deficient Mutants
To assess if the residual male transmission of seth1 and seth2 mutant pollen could be restored in the presence of a limited number of wild-type competitors, we performed a series of limited pollination experiments in which up to 25 pollen grains were placed onto stigmas of ms1-1 plants and the transmission of each mutant allele was determined. In these experiments, in which 120 to 220 F1 seedlings were scored, neither seth1 nor seth2 was transmitted through the male. Therefore, the failure of male transmission was not dependent on competition from wild-type pollen. These data indicate that the majority of GPI-deficient pollen grains that germinate are unable to reach the micropyle to achieve fertilization. This conclusion is supported by the impaired growth of seth mutant pollen tubes in vitro and the presence of short arrested pollen tubes on the stigma. Therefore, most mutant pollen tubes are affected severely and do not fail simply as a result of uncompetitive pollen tube growth.
SETH1 and SETH2 Are Expressed Widely in Sporophytic and Gametophytic Tissues
Our data demonstrate that GPI biosynthetic mutations result in gametophytic, male-specific effects on pollen germination and tube growth. However, GPI anchors are present in a variety of cells and tissues (Takos et al., 1997; Sherrier et al., 1999; Oxley and Bacic, 1999) such that widespread expression of GPI biosynthetic enzymes is expected. In this regard, we analyzed the expression of SETH1 and SETH2 in a range of tissue samples using reverse transcriptase–mediated (RT) PCR. SETH1 and SETH2 transcripts were detected in roots, stems, leaves, flowers, and isolated pollen (data not shown), demonstrating the expression of GPI biosynthetic genes in both sporophytic and gametophytic tissues.
Identification of Pollen-Expressed Genes Encoding GAPs
The requirement for GPI biosynthesis to support efficient pollen germination and tube growth suggests that one or more GAPs could be essential for these processes. To provide a genome-wide analysis of potential GAPs present in pollen, we used a transcriptomic approach. Bioinformatic predictions suggest that the Arabidopsis genome encodes 248 potential GAPs (Borner et al., 2003), but only a subset of these are likely to be expressed in pollen. We analyzed Affymetrix complete genome microarray hybridization data from two hybridizations with copy RNA prepared from mature pollen of two independent wild-type (Landsberg erecta) populations (Honys and Twell, 2003). Among the 212 predicted GAP genes embedded on the complete Arabidopsis genome array, we identified 41 genes that gave positive signals in both hybridization experiments (Table 2). The expression of all of these genes was confirmed by RT-PCR analysis of independent RNA samples from isolated pollen (Table 2). Most of the classes of GAPs defined by Borner et al. (2002)(2003) were represented.
Table 2.
Transcriptomic Analysis of GAP Genes Expressed in Mature Pollen
| AGI No. | GAP Family | Pollen Signal |
RT-PCR
|
||||
|---|---|---|---|---|---|---|---|
| Roots | Stems | Leaves | Flowers | Pollen | |||
| At1g24520 | Bcp1-like | 7371.3 | − | − | − | ± | +++ |
| At3g20865 | AG peptide (AtAGP40) | 6796.6 | − | − | − | ± | +++ |
| At3g57690 | AG peptide (AtAGP23) | 6720.2 | − | − | − | ± | ++ |
| At5g14380 | Classic AGPs (AtAGP6) | 6602.3 | − | − | − | − | +++ |
| At5g40730 | AG peptide (AtAGP24) | 5728.2 | ++ | ++ | + | ++ | +++ |
| At3g26110 | Unknown/hypothetical family 16 | 5559.6 | − | − | − | ± | ++ |
| At3g01700 | Classic AGPs (AtAGP11) | 5223.6 | − | − | − | + | +++ |
| At2g24450 a | Fasciclin-like (FLA3) | 5017.5 | − | − | − | + | +++ |
| At5g20230 a | Phytocyanin (stellacyanin-like) | 4779.8 | ± | + | ± | + | ++ |
| At2g20700 | Unknown/hypothetical family 4 | 4105.8 | − | − | ± | + | +++ |
| At5g58170 | Glycerophosphodiesterase-like (GPDL) | 3559.7 | − | − | ± | + | +++ |
| At5g58050 | Glycerophosphodiesterase-like (GPDL) | 3360 | − | − | − | − | + |
| At3g20580 | COBRA-like (COBL10) | 3211.1 | − | − | − | − | +++ |
| At4g28280 | Unknown/hypothetical family 4 | 2555.7 | − | − | − | − | +++ |
| At5g64790 a | β-1,3-Glucanase | 2223 | − | − | − | − | ++ |
| At5g53250 | AG peptide (AtAGP22) | 1706 | + | + | + | + | + |
| At5g36260 | Aspartylprotease | 1205.7 | − | − | − | ± | +++ |
| At4g08670 | Lipid transfer protein-like (LTPL) | 1086.8 | − | − | − | ++ | ++ |
| At1g65240 | Aspartylprotease | 1021.2 | ± | − | − | + | ++ |
| At4g27110 | COBRA-like (COBL11) | 906.4 | − | − | − | − | +++ |
| At1g55330 | AG peptide (AtAGP21) | 747.8 | + | + | ++ | ± | + |
| At1g18280 a | Lipid transfer protein-like (LTPL) | 730.5 | − | + | ± | − | ++ |
| At1g70170 | Metalloprotease | 455.4 | + | ± | + | ++ | +++ |
| At2g46330 | AG peptide (AtAGP16) | 226.4 | ++ | ++ | + | + | + |
| At3g27410 | Unknown/hypothetical family 6 | 208.2 | − | − | − | + | ++ |
| At3g12660 a | Fasciclin-like (FLA14) | 172.6 | + | + | + | + | + |
| At1g09790 | COBRA family (COBL6) | 140.2 | − | ± | − | − | ++ |
| At5g51480 | SKU5 family | 98.3 | − | + | ++ | ± | ++ |
| At3g16860 | COBRA-like (COBL8) | 90.5 | ± | + | ± | + | + |
| At1g05840 | Aspartylprotease | 87.7 | ++ | ++ | ++ | ++ | + |
| At1g56320 | Unknown/hypothetical family 11 | 79.5 | ± | ± | + | − | + |
| At5g49270 | COBRA-like (COBL9) | 71.7 | − | +++ | ++ | + | ++ |
| At4g28100 | Unknown/hypothetical family 5 | 64.8 | + | + | + | + | + |
| At3g61980 | Unknown/hypothetical family 19 | 61.5 | + | ± | ± | ± | + |
| At3g04010 | β-1,3-Glucanase | 50.8 | ± | ± | ± | ± | ± |
| At3g02740 | Aspartylprotease | 37.6 | + | ± | + | + | ++ |
| At4g16140 | Extensin-related | 31.3 | + | + | + | + | ++ |
| At5g60490 | Fasciclin-like (FLA12) | 25.6 | ± | ++ | ± | + | ++ |
| At3g58100 | β-1,3-Glucanase | 25.3 | − | + | ± | + | ++ |
| At1g77780 | β-1,3-Glucanase | 20.1 | ± | ± | + | − | ++ |
| At2g19440 | β-1,3-Glucanase | 17.9 | − | + | + | + | + |
| At1g64760 a | β-1,3-Glucanase | NR | − | 6 | 1 | 1 | 11 |
| At5g58480 a | β-1,3-Glucanase | NR | 6 | 1 | 1 | 1 | 111 |
| At3g18590 a | Phytocyanin (early nodulin-like) | NR | − | − | − | − | 111 |
| At1g48940 a | Phytocyanin (early nodulin-like) | NR | − | − | − | − | 111 |
| At3g07390 a | Auxin-induced protein (AIR12) | NR | 1 | 1 | 1 | 1 | 11 |
| At1g54860 a | Unknown/hypothetical family 1 | NR | 1 | 1 | − | − | 1 |
Affymetrix microarray hybridization data showing GAP genes expressed in mature pollen. Microarray signal intensities were normalized according to Honys and Twell (2003). RT-PCR analyses of all predicted GAP genes with positive microarray signals were performed in RNA samples from roots, stems, leaves, flowers, and mature pollen. Symbols indicate expression as follows: −, not detected; ±, weak; +, normal; ++, strong; +++, very strong.
GAP genes not represented (NR) on the microarray that were detected by proteomic analysis.
RT-PCR analysis of pollen and sporophytic tissues (root, stem, leaf, and flower) revealed that 15 of these 41 GAP mRNAs were detected specifically in flowers and pollen (Table 2). These tissue-specific transcripts encode a functionally divergent set of proteins: Bcp1-like, AG peptides, classic arabinogalactan proteins, fasciclin-like AGPs (FLAs), β-1,3-glucanases, aspartylproteases, glycerophosphodiesterase-like proteins, lipid transfer–like proteins, COBRA-like proteins, and unknown GAPs.
GAPs in Pollen
To directly identify GAPs present in pollen membranes, we used a proteomic approach relying on their altered partitioning in a two-phase system upon phosphatidylinositol-specific phospholipase C (Pi-PLC) cleavage. GAPs can be identified by a characteristic shift after anchor cleavage from the hydrophobic environment of the Triton X-114 detergent–rich phase into the aqueous phase (Sherrier et al., 1999). GAP-rich fractions were prepared by PLC treatment of hydrophobic proteins from mature Arabidopsis pollen membranes. To identify any abundant non-GAPs that contaminate the GAP-rich aqueous phase, we performed a control experiment without Pi-PLC. Proteins were separated by one-dimensional gel electrophoresis. Both treated and control lanes were cut into sections, and the proteins were digested with trypsin. The released peptides were analyzed by liquid chromatography–tandem mass spectrometry. Eleven GAPs were enriched specifically in the Pi-PLC–treated fraction (Table 3), including three β-1,3-glucanases, three phytocyanins, two FLAs, and one potential lipid transfer–like protein. Other GAPs are likely to have been below the detection limit of this analysis. All 11 GAPs were predicted to be GPI anchored by Borner et al. (2003).
Table 3.
Confirmed GAPs from Arabidopsis Pollen
| Number | Protein Family | MIPS Number | AGP | C-Terminus with Predicted Cleavage Site |
|---|---|---|---|---|
| 1 | β-1,3 Glucanase | At5g64790 | − | PVQIVS GSDDFRINFVFGRFVVFGLVLLGLLTVI |
| 2 | β-1,3 Glucanase | At1g64760 | − | IQIVAS SASSFSCSSYSLVVLIVWFLLSGMMF |
| 3 | β-1,3 Glucanase | At5g58480 | − | LDTSHS SSQTPNFFQSWPLLLLFLLSGLF |
| 4 | Phytocyanin (stellacyanin like) | At5g20230 | − | TTPAGN AASSLGGATFLVAFVSAVVALF |
| 5 | Phytocyanin (early nodulin like) | At3g18590 | − | AVQFSS SGFVVSAVLIVSVFGLV |
| 6 | Phytocyanin (early nodulin like) | At1g48940 | − | TSRFLG AGLVTISILVITVFSLV |
| 7 | Fasciclin like (FLA3) | At2g24450 | + | EAEPPS SASNTGLSFGAVLVLGFVASFVGF |
| 8 | Fasciclin like (FLA14) | At3g12660 | + | PSENAG SANGVSRNDSQPAFAFTLLMSFIWWFMARLR |
| 9 | Lipid transfer protein like (LTPL) | At1g18280 | − | TAKPTS SAPAINFGGLSFASAVVATLFF |
| 10 | Auxin induced protein (AIR12) | At3g07390 | + | AGGPGN AGSLTRNVNFGVNLGILVLLGSIFIF |
| 11 | Unknown/Hypothetical Family 1 | At1g54860 | − | FTAGVA AGKATSVRVMAGLGLMGLLFSCLVLF |
Proteins were separated by one-dimensional SDS-PAGE. Proteins sensitive to Pi-PLC were identified by liquid chromatography–tandem mass spectrometry. Only identifications with >95% confidence based on MASCOT (www.matrixscience.com) scores were considered significant. The most likely cleavage sites (Udenfriend and Kodukula, 1995) are indicated (boldface). The hydrophobic domain of each C-terminal signal peptide is underlined. Confirmations have been submitted to the Munich Information Center for Protein Sequences.
RT-PCR analysis (Table 2) confirmed that all of the 11 corresponding genes were expressed in pollen, 7 of them in a tissue-specific manner. At5g64790, At3g18590, At1g48940, and At2g24450 were expressed preferentially in pollen, whereas the expression of At1g64760, At1g18280, and At1g54860 was restricted to pollen and certain sporophytic tissues.
DISCUSSION
GPI modification of proteins occurs in most eukaryotic systems and appears to be essential for the modulation of cell surface properties. In plants, GAPs fulfill a number of potential functions, although the significance of GPI anchoring and the biological roles of GAPs remain poorly understood. We identified and characterized four insertional mutations that disrupt two probable subunits of the GPI-GnT complex, PIG-C (seth1) and PIG-A (seth2). Heterozygous GPI-GnT complex mutations in Arabidopsis have no effect on sporophytic development and megagametogenesis but show gametophytic, male-specific defects in fertility. Mutant pollen is viable, but the majority of GPI-deficient pollen grains fail during either germination or early tube growth. Our results suggest that GPI anchoring is required during both phases of post-pollination development.
Recent studies in mice also have demonstrated a reproductive role for GPI anchoring, but in female fertility. Conditional PIG-A knockout female mice are infertile, and eggs recovered after mating remain unfertilized, suggesting that GAPs on the egg surface play a role in gamete fusion (Alfieri et al., 2003). Our results do not support a role for GPI anchoring in gamete fusion in Arabidopsis. However, because plant egg cells are not physically independent, essential GAPs in GPI-deficient embryo sacs could be supplied from surrounding sporophytic cells.
Although highly penetrant, GPI-GnT complex mutations occasionally were transmitted through pollen. Therefore, a low percentage of mutant pollen grains were able to germinate and develop functional pollen tubes. In vivo germination assays indicated that 26% of pollen grains carrying the seth2 mutant allele were able to germinate, but genetic analysis revealed only 1.6% male transmission, suggesting that GPIs also are required during pollen germination and pollen tube elongation. Indeed, GPI-deficient pollen tubes grown in vitro were shorter that their wild-type counterparts, and most showed high levels of abnormal callose deposition. In vivo analysis showed that only 9.2% of seth2 pollen grains (4.6% of the total population) were able to develop sufficiently long pollen tubes to allow cytoplasmic translocation. Together, these data suggest that 74% of seth2 pollen fail to germinate and another 16.8% develop only short pollen tubes.
The residual transmission of GPI-deficient mutations might be explained by the existence of a pool of GAPs, GPIs, or GPI precursors associated with the endoplasmic reticulum and inherited through meiosis. Saccharomyces cerevisiae GPI-deficient haploid ascospores are able to germinate and complete up to four cell divisions (Leidich et al., 1995). Moreover, GAPs added to animal cell cultures can be incorporated into surface membranes and exert native functions (Premkumar et al., 2001); therefore, it is possible that pollen tubes may acquire GPI from surrounding pistil tissues. On the other hand, the significant number (17 of 47) of pollen-expressed GAP genes that were not expressed detectably in sporophytic tissues suggests that many GAPs are not inherited through meiosis. However, it remains possible that some GAPs could be supplied from within the anther locule or by the papillar cells and transmitting tissue during pollen tube growth.
The total number of Arabidopsis proteins predicted to be attached to the cell surface by GPI anchoring is 248 (Borner et al., 2002, 2003). We identified 41 GAP genes expressed in pollen among the 212 predicted GAP genes embedded on the Arabidopsis genome array. Six additional GAP genes were identified by proteomic analysis of mature pollen. Therefore, our data demonstrate that at least 47 potential Arabidopsis GAPs are expressed in pollen. Thus, the failure of pollen germination and/or tube growth in seth1 and seth2 mutants could result from the absence of one or more of these cell surface proteins.
GAPs are targeted to the cell surface in a polarized manner in many organisms. There is increasing evidence that many GAPs in animal and yeast cells can be clustered into sphingolipid- and sterol-enriched membrane microdomains, known as lipid rafts (reviewed by Muniz and Riezman, 2000), and there are indications that such membrane microdomains exist in higher plants (Peskan et al., 2000; Willemsen et al., 2003; G.H.H. Borner and P. Dupree, unpublished data). As demonstrated in other polarized cell types (Ikonen, 2001; Bagnat and Simons, 2002; Rodriguez-Peña et al., 2002; Tsui-Pierchala et al., 2002), microdomains could be crucial for polarized cell processes, such as pollen germination and tip growth. In mammalian cells, GAPs can be endocytosed via a clathrin-independent pathway to specialized endosomes or the Golgi apparatus before being recycled back to the cell surface (Fivaz et al., 2002; Nichols, 2002; Sabharanjak et al., 2002). We speculate that GAPs could be targeted to the tip region during pollen germination and tube growth, perhaps by lipid raft–dependent processes. Moreover, endocytic recycling of GAPs from the pollen tube flanks to the apex could maintain the localization of GAPs within the apical region. Given our identification of GPI-anchored β-1,3-glucanases in pollen membranes, we hypothesize that their potential tip localization could antagonize pollen-expressed callose synthases (Doblin et al., 2001), restricting callose synthesis to the pollen tube flanks. This notion is consistent with the abnormally high levels of callose present in seth1 and seth2 pollen tubes and the overaccumulation of callose in nongerminated seth1 and seth2 pollen. These phenotypes could result from disturbances in the synthesis and/or mislocalization of gametophytic GPI-anchored β-1,3-glucanases. In addition, GPI anchoring also could help maintain the polar distribution of other GAPs that may function in signaling events during pollen germination and at the pollen tube tip.
It may be significant that all four Arabidopsis GAPs characterized to date have roles in directional growth or cell expansion and/or influence the properties of the cell wall. COBRA is required for polarized longitudinal expansion in the root, which Schindelman et al. (2001) proposed might result from its recruitment of cellulose-synthesizing complexes at the cell surface. SKU5 acts on the control of directional growth in roots, but its mode of action remains unknown (Sedbrook et al., 2002). A mutation in PMR6, which encodes a pectate lyase, alters cell wall organization and pectin content (Vogel et al., 2002). Mutations in SOS5/AtFLA4 result in abnormal root cell expansion and growth arrest under salt stress (Shi et al., 2003). It is difficult to know at this time which GAPs are essential for pollen germination and tube growth and what their mode of action is. We suggest that the severe defect in pollen tube germination may result from the disruption of protein targeting and/or the absence of more than one of these pollen GPI-anchored cell surface proteins. Analysis of knockout mutant lines will allow the role of individual proteins to be investigated.
GPI deficiency might differentially affect the fate of GAP precursors, but it is likely to prevent their correct targeting to the plasma membrane. For example, in GPI-deficient mutants generated by the heterologous expression of Trypanosoma brucei GPI-PLC, of four proteins that normally are GPI anchored in Trypanosoma cruzi, two were secreted prematurely and the other two were degraded intracellularly (Garg et al., 1997). GAPs that are transferred incompletely to the GPI anchor in yeast are retained in the endoplasmic reticulum and degraded (Horvath et al., 1994). The fate of GPI precursor proteins in pollen GPI-deficient mutants could be addressed using either specific antibodies or GPI precursor protein:green fluorescent protein fusions (Sedbrook et al., 2002).
Moreover, the potential importance of free GPIs should not be neglected. Although we are unaware of any evidence from plants, free GPIs are present at the surfaces of mammalian (Singh et al., 1996) and protozoal (Ilgoutz et al., 1999) cells and may be involved in the organization of microdomains and membrane fluidity (Muniz and Riezman, 2000). At least in Leishmania, free GPIs appear to play an important role in cell elongation (Ilgoutz et al., 1999). However, there are conflicting data regarding the importance of free GPIs compared with GPI-linked proteins. GAPs were found not to be required for lymphocyte development in mice, but were essential for normal lymphocyte function and maintenance of a normal peripheral lymphoid compartment (Bessler et al., 2002). Similarly, GAPs are not essential to Leishmania for survival within mammalian host cells (Hilley et al., 2000), but parasite growth depends of the availability of free GPIs (Ilgoutz et al., 1999). Furthermore, transamidation-deficient mutants in yeast accumulate complete GPI lipids as well as GPI precursor proteins and show cell growth arrest (Meyer et al., 2000). Mutational analysis of GPI biosynthesis targeting the transamidation step might provide additional insights into the importance of free GPIs and GAPs for pollen germination and tube growth, respectively.
The diversity of GAPs expressed in pollen highlights the complexity of the mechanisms underlying post-pollination events. Pollen GAP protein homologies suggest functions associated with the regulation of the structural properties and synthesis of the pollen tube wall and signaling at the pollen tube surface. GPI-deficient pollen, which is viable until germination, represents a valuable in vivo system that allows the stepwise dissection of GPI biosynthesis and the functional analysis of GPI-anchored cell surface proteins in plants.
METHODS
Mutant Lines and Growth Conditions
DsE and DsG lines in the Landsberg erecta ecotype were generated by U. Grossniklaus and co-workers at Cold Spring Harbor Laboratory (Cold Spring Harbor, NY) as described (Sundaresan et al., 1995; Moore et al., 1997). seth1-1 was isolated from a genetic screen of 3616 Ds lines based on segregation ratio distortion (E. Lalanne et al., unpublished data). Thermal asymmetric interlaced PCR was performed according to Liu et al. (1995) with minor modifications. Ds-flanking sequences were amplified using three GUS-specific nested primers (GUS-1, 5′-CGTAATGAGTGACCGCATCG-3′; GUS-2, 5′-GACGTTGCCCGCATAATTAC-3′; GUS-3, 5′-GATCCAGACTGAATGCCCAC-3′) combined with the AD1, AD2, or AD3 degenerate primers (Liu et al., 1995). The Ds-flanking DNA junction was confirmed using Ds5-1 (Grossniklaus et al., 1998) and a gene-specific primer (5′-ATCATGCAAGAAGAGATGAAG-3′). seth1-2 (Garlic_674B03) and seth1-3 (Garlic_165E01) were obtained from Syngenta. seth2 (SALK_039500) was generated by the Salk Institute. Insertion mutant information was obtained from the SIGnAL World Wide Web site at http://signal.salk.edu.
seth1-1 and seth2 seeds were sterilized in a drop of 95% ethanol. Ethanol was allowed to evaporate overnight. Sterilized seeds were plated onto kanamycin (50 ng/L)–supplemented medium (0.5× Murashige and Skoog [1962] salts [Sigma] and 0.8% agar, pH 5.8). After 2 days at 4°C, plates were incubated under continuous light at 21°C. seth1-2 and seth1-3 seeds were plated on a 3:1 compost:sand mix watered with 0.83 μg/L glufosinate (ammonium) (Final; Hoechst, Marseille, France). After 2 days at 4°C, the seeds were grown under greenhouse conditions with supplemental lighting (16 h of light at 22°C). Antibiotic or herbicide phenotypes were scored after 10 days, and resistant seedlings were transferred to 3-cm2 pots containing a 3:1 compost:sand mix and grown under greenhouse conditions.
Genetic Transmission through Male and Female Gametes
Genetic transmission of mutations through the male and female gametes was determined by reciprocal test crosses of heterozygous mutants and the wild type (Landsberg erecta) according to Howden et al. (1998). The transmission efficiency of the mutant allele through each gamete was calculated according to Howden et al. (1998). Limited pollinations were performed on stigmas of ms1-1 male-sterile plants (Wilson et al., 2001). Up to 25 pollen grains were deposited onto ms1-1 stigmas using a dissecting needle and a dissecting microscope (Stemi SV-6 stereomicroscope; Zeiss, Jena, Germany).
Cytological and Phenotypic Analyses of Pollen
Mature pollen grains were incubated in 4′,6-diamidino-2-phenylindole staining solution and observed using light and epifluorescence microscopy as described by Park et al. (1998). Alexander and aniline blue staining of mature pollen grains were performed as described by Alexander (1969) and Park and Twell (2001), respectively. Cellulose or pectin staining was performed using 0.1% calcofluor (Fluorescent Brightener 28; Sigma) or 0.01% ruthenium red (Sigma), respectively. For each mutant line, >4000 pollen grains from 10 independent plants were examined.
In vitro germination assays were performed as follows. Individual open flowers were collected in microtiter plates (TC microwell 96F; Nucleon Biosciences, Glasgow, Scotland) containing 50 μL of germination medium (Derksen et al., 2002) per well. Plates were sealed and incubated overnight at 22°C under continuous light. More than 3000 pollen grains per line were scored by direct observation of plates using a Zeiss Axiovert inverted microscope. NIH Image version 1.6 was used to measure pollen tube lengths from captured images.
To determine in vivo germination efficiency, excised ms1-1 stigmas were cut at the base and inserted vertically into solid agar (1% in water) in a 9-cm Petri dish. Limited pollinations were performed as described above. Stigmas were transferred on a microscope slide and stained with aniline blue solution at 2 h after pollination or with Alexander staining at 4 h after pollination. More than 400 pollen grains deposited on 20 stigmas were analyzed per line.
RNA Extraction and Affymetrix Complete Genome Microarray Hybridization
RNA extraction, microarray hybridization, and reverse transcriptase–mediated PCR analyses were performed as described by Honys and Twell (2003). Oligonucleotide sequences and annealing temperatures are provided in the supplemental data online.
Biochemical Fractionation and Preparation of GPI-Anchored Proteins
Pollen grains were resuspended in 3 volumes of 5% sucrose/TE buffer (100 mM Tris and 1 mM EDTA, pH 8.0) and homogenized in a Dounce glass homogenizer. After centrifugation for 10 min at 1614g, the supernatant was collected. The insoluble pellet was resuspended in 1 volume of 5% sucrose/TE buffer and reextracted by sonication (Ultrasonic Processor XL; Heat Systems, Farmingdale, NY) for 4 × 15 s at level 5. The extract was centrifuged at 1614g for 20 min. The supernatant was pooled with that obtained after homogenization, loaded onto a sucrose step gradient 10%/48%, and centrifuged for 3 h at 100,000g. The membrane fraction was harvested at the 10%/48% interphase and diluted fivefold in TE buffer. Membranes were pelleted at 100,000g for 2 h. Membrane pellets were resuspended in TBS buffer (10 mM Tris and 150 mM NaCl, pH 7.4, at 37°C), frozen in liquid nitrogen, and stored at −80°C in aliquots. GPI-anchored proteins were prepared as described by Borner et al. (2003) using phosphatidylinositol-specific phospholipase C (Sigma, Poole, UK) in conjunction with Triton X-114 phase partitioning.
Electrophoretic Analysis and Mass Spectrometry
GPI-anchored proteins were analyzed by one-dimensional SDS-PAGE and identified by liquid chromatography–tandem mass spectrometry as described by Borner et al. (2003).
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact David Twell, twe@le.ac.uk.
Supplementary Material
Acknowledgments
We thank the Salk Institute Genomic Analysis Laboratory for providing sequence-indexed Arabidopsis T-DNA insertion mutants. We thank the Torrey Mesa Research Institute (Syngenta) for providing insertion lines from the SAIL collection, and James Moore, Wendy Gagliano, and Jean-Phillippe Vielle Calzada for help in generating Ds insertion lines. Seeds of ms1-1 and SALK T-DNA mutants were provided by the Nottingham Arabidopsis Stock Centre. Funding for the SIGnAL indexed insertion mutant collection was provided by the National Science Foundation. This work was made possible by research grants from the Biotechnology and Biological Sciences Research Council (BBSRC), the BBSRC Investigating Gene Function Initiative, and services provided through the GARNet transcriptomic and proteomic facilities. D.H. was supported through a Royal Society/North Atlantic Treaty Organization fellowship and Academy of Sciences of the Czech Republic Grant IAA5038207. G.H.H.B. received a BBSRC research studentship and a scholarship from the Studienstiftung des Deutschen Volkes. U.G. was supported by the Cold Spring Harbor Laboratory President's Council, the European Molecular Biology Organization, the Human Frontier Science Program, and the Kanton of Zürich.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.014407.
Footnotes
Online version contains Web-only data.
References
- Alexander, M.P. (1969). Differential staining of aborted and nonaborted pollen. Stain Technol. 44, 117–122. [DOI] [PubMed] [Google Scholar]
- Alfieri, J.A., Martin, A.D., Takeda, J., Kondoh, G., Myles, D.G., and Primakoff, P. (2003). Infertility in female mice with an oocyte-specific knockout of GPI-anchored proteins. J. Cell Sci. 116, 2149–2155. [DOI] [PubMed] [Google Scholar]
- Bagnat, M., and Simons, K. (2002). Cell surface polarization during yeast mating. Proc. Natl. Acad. Sci. USA 99, 14183–14188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benachour, A., Sipos, G., Flury, I., Reggiori, F., Canivenc-Gansel, E., Vionnet, C., Conzelmann, A., and Benghezal, M. (1999). Deletion of GPI7, a yeast gene required for addition of a side chain to the glycosylphosphatidylinositol (GPI) core structure, affects GPI protein transport, remodeling, and cell wall integrity. J. Biol. Chem. 274, 15251–15261. [DOI] [PubMed] [Google Scholar]
- Bessler, M., Rosti, V., Peng, Y., Cattoretti, G., Notaro, R., Ohsako, S., Elkon, K.B., and Luzzatto, L. (2002). Glycosylphosphatidylinositol-linked proteins are required for maintenance of a normal peripheral lymphoid compartment but not for lymphocyte development. Eur. J. Immunol. 32, 2607–2616. [DOI] [PubMed] [Google Scholar]
- Borner, G.H.H., Lilley, K.S., Stevens, T.J., and Dupree, P. (2003). Identification of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: A proteomic and genomic analysis. Plant Physiol. 132, 568–577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Borner, G.H.H., Sherrier, D.J., Stevens, T.J., Arkin, I.T., and Dupree, P. (2002). Prediction of glycosylphosphatidylinositol-anchored proteins in Arabidopsis: A genomic analysis. Plant Physiol. 129, 486–499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Derksen, J., Knuiman, B., Hoedemaekers, K., Guyon, A., Bonhomme, S., and Pierson, E.S. (2002). Growth and cellular organization of Arabidopsis pollen tubes in vitro. Sex. Plant Reprod. 15, 133–139. [Google Scholar]
- Doblin, M.S., De Melis, L., Newbigin, E., Bacic, A., and Read, S.M. (2001). Pollen tubes of Nicotiana alata express two genes from different β-glucan synthase families. Plant Physiol. 125, 2040–2052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fivaz, M., Vilbois, F., Thurnheer, S., Pasquali, C., Abrami, L., Bickel, P.E., Parton, R.G., and van der Goot, F.G. (2002). Differential sorting and fate of endocytosed GPI-anchored proteins. EMBO J. 21, 3989–4000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Garg, N., Tarleton, R.L., and Mensa-Wilmot, K. (1997). Proteins with glycosylphosphatidylinositol (GPI) signal sequences have divergent fates during a GPI deficiency: GPIs are essential for nuclear division in Trypanosoma cruzi. J. Biol. Chem. 272, 12482–12491. [DOI] [PubMed] [Google Scholar]
- Gaynor, E.C., Mondesert, G., Grimme, S.J., Reed, S.I., Orlean, P., and Emr, S.D. (1999). MCD4 encodes a conserved endoplasmic reticulum membrane protein essential for glycosylphosphatidylinositol anchor synthesis in yeast. Mol. Biol. Cell 10, 627–648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grossniklaus, U., Vielle-Calzada, J.-P., Hoeppner, M.A., and Gagliano, W.B. (1998). Maternal control of embryogenesis by MEDEA, a Polycomb-group gene in Arabidopsis. Science 280, 446–450. [DOI] [PubMed] [Google Scholar]
- Hilley, J.D., Zawadzki, J.L., McConville, M.J., Coombs, G.H., and Mottram, J.C. (2000). Leishmania mexicana mutants lacking glycosylphosphatidylinositol (GPI):protein transamidase provide insights into the biosynthesis and functions of GPI-anchored proteins. Mol. Biol. Cell 11, 1183–1195. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hong, Y., Maeda, Y., Watanabe, R., Ohishi, K., Mishkind, M., Riezman, H., and Kinoshita, T. (1999). Pig-n, a mammalian homologue of yeast Mcd4p, is involved in transferring phosphoethanolamine to the first mannose of the glycosylphosphatidylinositol. J. Biol. Chem. 274, 35099–35106. [DOI] [PubMed] [Google Scholar]
- Honys, D., and Twell, D. (2003). Comparative analysis of the Arabidopsis pollen transcriptome. Plant Physiol. 132, 640–652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horvath, A., Sütterlin, C., Manning-Krieg, U., Movva, N.R., and Riezman, H. (1994). Ceramide synthesis enhances transport of GPI-anchored proteins to the Golgi apparatus in yeast. EMBO J. 13, 3687–3695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Howden, R., Park, S.K., Moore, J.M., Orme, J., Grossniklaus, U., and Twell, D. (1998). Selection of T-DNA-tagged male and female gametophytic mutants by segregation distortion in Arabidopsis. Genetics 149, 621–631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikezawa, H. (2002). Glycosylphosphatidylinositol (GPI)-anchored proteins. Biol. Pharm. Bull. 25, 409–417. [DOI] [PubMed] [Google Scholar]
- Ikonen, E. (2001). Roles of lipid rafts in membrane transport. Curr. Opin. Cell Biol. 13, 470–477. [DOI] [PubMed] [Google Scholar]
- Ilgoutz, S.C., Zawadzki, J.L., Ralton, J.E., and McConville, M.J. (1999). Evidence that free GPI glycolipids are essential for growth of Leishmania mexicana. EMBO J. 18, 2746–2755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imbach, T., et al. (2000). Deficiency of dolichol-phosphate-mannose synthase-1 causes congenital disorder of glycosylation type Ie. J. Clin. Invest. 105, 233–239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inoue, N., Watanabe, R., Takeda, J., and Kinoshita, T. (1996). PIG-C, one of the three human genes involved in the first step of glycosylphosphatidylinositol biosynthesis, is a homologue of Saccharomyces cerevisiae GPI2. Biochem. Biophys. Res. Commun. 226, 193–199. [DOI] [PubMed] [Google Scholar]
- Johnson, S.A., and McCormick, S. (2001). Pollen germinates precociously in the anthers of raring-to-go, an Arabidopsis gametophytic mutant. Plant Physiol. 126, 685–695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leidich, S.D., Kostova, Z., Latek, R.R., Costello, L.C., Drapp, D.A., Gray, W., Fassler, J.S., and Orlean, P. (1995). Temperature-sensitive yeast GPI anchoring mutants gpi2 and gpi3 are defective in the synthesis of N-acetylglucosaminyl phosphatidylinositol: Cloning of the GPI2 gene. J. Biol. Chem. 270, 13029–13035. [DOI] [PubMed] [Google Scholar]
- Liu, Y.G., Mitsukawa, N., Oosumi, T., and Whittier, R.F. (1995). Efficient isolation and mapping of Arabidopsis thaliana T-DNA insert junctions by thermal asymmetric interlaced PCR. Plant J. 8, 457–463. [DOI] [PubMed] [Google Scholar]
- Meyer, U., Benghezal, M., Imhof, I., and Conzelmann, A. (2000). Active site determination of Gpi8p, a caspase-related enzyme required for glycosylphosphatidylinositol anchor addition to proteins. Biochemistry 39, 3461–3471. [DOI] [PubMed] [Google Scholar]
- Miyata, T., Takeda, J., Iida, Y., Yamada, N., Inoue, N., Takahashi, M., Maeda, K., Kitani, T., and Kinoshita, T. (1993). The cloning of PIG-A, a component in the early step of GPI-anchor biosynthesis. Science 259, 1318–1320. [DOI] [PubMed] [Google Scholar]
- Moore, J.M., Vielle Calzada, J.-P., Gagliano, W., and Grossniklaus, U. (1997). Genetic characterization of hadad, a mutant disrupting megagametogenesis in Arabidopsis thaliana. Cold Spring Harbor Symp. Quant. Biol. 62, 35–47. [PubMed] [Google Scholar]
- Muniz, M., and Riezman, H. (2000). Related intracellular transport of GPI-anchored proteins. EMBO J. 19, 10–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nichols, B.J. (2002). A distinct class of endosome mediates clathrin-independent endocytosis to the Golgi complex. Nat. Cell Biol. 5, 374–378. [DOI] [PubMed] [Google Scholar]
- Oxley, D., and Bacic, A. (1999). Structure of the glycosylphosphatidylinositol anchor of an arabinogalactan protein from Pyrus communis suspension-cultured cells. Proc. Natl. Acad. Sci. USA 96, 14246–14251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park, S.K., Howden, R., and Twell, D. (1998). The Arabidopsis thaliana gametophytic mutation gemini pollen 1 disrupts microspore polarity, division asymmetry and pollen cell fate. Development 125, 3789–3799. [DOI] [PubMed] [Google Scholar]
- Park, S.K., and Twell, D. (2001). Novel patterns of ectopic cell plate growth and lipid body distribution in the Arabidopsis gemini pollen1 mutant. Plant Physiol. 126, 899–909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peskan, T., Westermann, M., and Oelmuller, R. (2000). Identification of low-density Triton X-100-insoluble plasma membrane microdomains in higher plants. Eur. J. Biochem. 267, 6989–6995. [DOI] [PubMed] [Google Scholar]
- Premkumar, D.R., Fukuoka, Y., Sevlever, D., Brunschwig, E., Rosenberry, T.L., Tykocinski, M.L., and Medof, M.E. (2001). Properties of exogenously added GPI-anchored proteins following their incorporation into cells. J. Cell. Biochem. 82, 234–245. [DOI] [PubMed] [Google Scholar]
- Preuss, D., Rhee, S.Y., and Davis, R.W. (1994). Tetrad analysis possible in Arabidopsis with mutation of the QUARTET (QRT) genes. Science 264, 1458–1460. [DOI] [PubMed] [Google Scholar]
- Rodriguez-Peña, J.M., Rodriguez, C., Alvarez, A., Nombela, C., and Arroyo, J. (2002). Mechanisms for targeting of the Saccharomyces cerevisiae GPI-anchored cell wall protein Crh2p to polarised growth sites. J. Cell Sci. 115, 2549–2558. [DOI] [PubMed] [Google Scholar]
- Rosti, V. (2000). The molecular basis of paroxysmal nocturnal hemoglobinuria. Haematologica 85, 82–87. [PubMed] [Google Scholar]
- Roudier, F., Schindelman, G., DeSalle, R., and Benfey, P.N. (2002). The COBRA family of putative GPI-anchored proteins in Arabidopsis: A new fellowship in expansion. Plant Physiol. 130, 538–548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabharanjak, S., Sharma, P., Parton, R.G., and Mayor, S. (2002). GPI-anchored proteins are delivered to recycling endosomes via a distinct cdc42-regulated, clathrin-independent pinocytic pathway. Dev. Cell 4, 411–423. [DOI] [PubMed] [Google Scholar]
- Schindelman, G., Morikami, A., Jung, J., Baskin, T.I., Carpita, N.C., Derbyshire, P., McCann, M.C., and Benfey, P.N. (2001). COBRA encodes a putative GPI-anchored protein, which is polarly localized and necessary for oriented cell expansion in Arabidopsis. Genes Dev. 15, 1115–1127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schultz, C.J., Johnson, K.L., Currie, G., and Bacic, A. (2000). The classical arabinogalactan protein gene family of Arabidopsis. Plant Cell 12, 1751–1768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedbrook, J.C., Carroll, K.L., Hung, K.F., Masson, P.H., and Somerville, C.R. (2002). The Arabidopsis SKU5 gene encodes an extracellular glycosyl phosphatidylinositol–anchored glycoprotein involved in directional root growth. Plant Cell 14, 1635–1648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sessions, A., et al. (2002). A high-throughput Arabidopsis reverse genetics system. Plant Cell 14, 2985–2994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sherrier, D.J., Prime, T.A., and Dupree, P. (1999). Glycosylphosphatidylinositol-anchored cell-surface proteins from Arabidopsis. Electrophoresis 20, 2027–2035. [DOI] [PubMed] [Google Scholar]
- Shi, H., Kim, Y., Guo, Y., Stevenson, B., and Zhu, J.K. (2003). The Arabidopsis SOS5 locus encodes a putative cell surface adhesion protein and is required for normal cell expansion. Plant Cell 15, 19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh, N., Liang, L.N., Tykocinski, M.L., and Tartakoff, A.M. (1996). A novel class of cell surface glycolipids of mammalian cells: Free glycosyl phosphatidylinositols. J. Biol. Chem. 271, 12879–12884. [DOI] [PubMed] [Google Scholar]
- Sundaresan, V., Springer, P., Volpe, T., Haward, S., Jones, J.D., Dean, C., Ma, H., and Martienssen, R. (1995). Patterns of gene action in plant development revealed by enhancer trap and gene trap transposable elements. Genes Dev. 9, 1797–1810. [DOI] [PubMed] [Google Scholar]
- Takos, A.M., Dry, I.B., and Soole, K.L. (1997). Detection of glycosyl-phosphatidylinositol-anchored proteins on the surface of Nicotiana tabacum protoplasts. FEBS Lett. 405, 1–4. [DOI] [PubMed] [Google Scholar]
- Tsui-Pierchala, B.A., Encinas, M., Milbrandt, J., and Johnson, E.M. (2002). Lipid rafts in neuronal signaling and function. Trends Neurosci. 25, 412–417. [DOI] [PubMed] [Google Scholar]
- Udenfriend, S., and Kodukula, K. (1995). How glycosylphosphatidylinositol-anchored membrane proteins are made. Annu. Rev. Biochem. 64, 563–591. [DOI] [PubMed] [Google Scholar]
- Willemsen, V., Friml, J., Grebe, M., Van Den Toorn, A., Palme, K., and Scheres, B. (2003). Cell polarity and PIN protein positioning in Arabidopsis require STEROL METHYLTRANSFERASE1 function. Plant Cell 15, 612–625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson, Z.A., Morroll, S.M., Dawson, J., Swarup, R., and Tighe, P.J. (2001). The Arabidopsis MALE STERILITY1 (MS1) gene is a transcriptional regulator of male gametogenesis, with homology to the PHD-finger family of transcription factors. Plant J. 28, 27–39. [DOI] [PubMed] [Google Scholar]
- Vogel, J.P., Raab, T.K., Schiff, C., and Somerville, S.C. (2002). PMR6, a pectate lyase–like gene required for powdery mildew susceptibility in Arabidopsis. Plant Cell 14, 2095–2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.