Abstract
A novel insight into Arabidopsis mitochondrial function was revealed from a large experimental proteome derived by liquid chromatography–tandem mass spectrometry. Within the experimental set of 416 identified proteins, a significant number of low-abundance proteins involved in DNA synthesis, transcriptional regulation, protein complex assembly, and cellular signaling were discovered. Nearly 20% of the experimentally identified proteins are of unknown function, suggesting a wealth of undiscovered mitochondrial functions in plants. Only approximately half of the experimental set is predicted to be mitochondrial by targeting prediction programs, allowing an assessment of the benefits and limitations of these programs in determining plant mitochondrial proteomes. Maps of putative orthology networks between yeast, human, and Arabidopsis mitochondrial proteomes and the Rickettsia prowazekii proteome provide detailed insights into the divergence of the plant mitochondrial proteome from those of other eukaryotes. These show a clear set of putative cross-species orthologs in the core metabolic functions of mitochondria, whereas considerable diversity exists in many signaling and regulatory functions.
INTRODUCTION
Mitochondria perform a variety of biochemical functions within the eukaryotic cell. Their primary roles are the oxidation of organic acids via the tricarboxylic acid cycle and the synthesis of ATP coupled to the transfer of electrons from reduced NAD+ to oxygen via the electron transport chain. However, in plants, mitochondria perform many important secondary functions, such as the synthesis of nucleotides, amino acids, lipids, and vitamins (Rebeille et al., 1997; Bartoli et al., 2000; Gueguen et al., 2000). They contain their own genome (Gray et al., 1999), undertake transcription and translation (Brennicke et al., 1999), actively import proteins and metabolites from the cytosol (Braun and Schmitz, 1999; Laloi, 1999; Lister et al., 2003), influence programmed cell death (Balk et al., 1999; Balk and Leaver, 2001), and respond to cellular signals such as oxidative stress (Møller, 2001; Sweetlove et al., 2002). Given these roles, it is essential that we understand the full gamut of mitochondrial function in plants and the mechanisms that govern their biogenesis, especially the way in which mitochondrial activity is perceived by the nucleus and coordinated with the rest of cellular function. A major limitation in such analyses has been a lack of precise information about the protein components of mitochondria potentially involved in these processes. Once these are known, reverse genetics has the capacity to dissect these pathways and to provide genetic engineering of mitochondrial function in plants.
Mitochondria are considered to have arisen from an endosymbiotic event between bacterial and eukaryotic ancestors. Since that time, a considerable reduction in the coding capacity of the mitochondrion has occurred through the transfer of most endosymbiont genes to the nucleus. The genomes of current mitochondria code a very limited subset of organelle proteins, with the majority encoded in the nucleus and targeted to mitochondria through recently evolved import mechanisms (for review, see Gray et al., 1999). Interestingly, although a single endosymbiotic event probably brought the organelle structures we call mitochondria into cells, a whole series of events that yielded mitochondria, plastids, and peroxisomes likely contributed to the current mitochondrial proteome (Adams et al., 2000, 2002; Gray et al., 2001; Richly et al., 2003).
Many of the nucleus-encoded proteins of the mitochondrial proteome are targeted to the mitochondria through signal sequences that usually reside in the N-terminal portion of the protein. In theory, the subcellular location of a protein can be predicted through commonly identified characteristics in these signal sequences. Several bioinformatic methods have been developed to predict the subcellular localization of a protein using both defined characteristics and machine-learning techniques (reviewed by Feng, 2001). This has led to a number of publicly available programs that can be used to predict protein subcellular localization. MitoProt II (Claros and Vincens, 1996), PSORT (Nakai and Horton, 1999), and iPSORT (Bannai et al., 2002) use predefined parameters of signal peptides to predict subcellular localization. The programs TargetP (Emanuelsson et al., 2000), Predotar (http://www.inra.fr/predotar/), and SubLoc (Hua and Sun, 2001) use machine-learning techniques for subcellular predictions. Recently, using a novel combination of comparative genomics and targeting prediction programs, Richly et al. (2003) analyzed 10 different eukaryotic genomes and proposed putative mitochondrial proteomes for each. The veracity of these various prediction sets is unclear at present because of the lack of substantial experimental data for comparisons.
Experimental approaches also have sought to define the size of the mitochondrial proteome. Two-dimensional electrophoresis studies indicate that mitochondrial samples from plants can be resolved into 500 to 1500 protein spots (Kruft et al., 2001; Millar et al., 2001b; Bardel et al., 2002). Similar numbers are noted in mammalian and yeast mitochondria (Rabilloud et al., 1998; Lopez et al., 2000; Scheffler et al., 2001). However, because a single protein often can appear as multiple protein spots by two-dimensional PAGE, the nonredundant set of proteins in these gels is considerably less. The completion of several sequencing projects also has enabled large-scale analyses and estimations of mitochondrial localization to be undertaken using other methods. Using phylogenetic profiles, Marcotte et al. (2000) estimated that there are ∼660 nucleus-encoded mitochondrial proteins in Caenorhabditis elegans and that the yeast nuclear genome encodes∼630 mitochondrial proteins. A proteome-scale analysis in yeast using random and directed epitope-tagging techniques to determine protein subcellular distribution estimated that 14% of the yeast proteome (∼850 proteins) is likely to be localized in mitochondria (Kumar et al., 2002). Currently, ∼580 yeast open reading frames at the Comprehensive Yeast Genome Database (http://mips.gsf.de/genre/proj/yeast/search_index.jsp) have been designated definitively as coding mitochondrial proteins. Furthermore, Schon (2001) has published a list of ∼550 yeast mitochondrial proteins assembled from identifications determined through a variety of experimental systems. More recently, an analysis of mitochondrial proteomes from human heart and yeast using mass spectrometric techniques identified 615 and 750 proteins, respectively (Sickmann et al., 2003; Taylor et al., 2003b).
Initial investigations of the components of the mitochondrial proteomes of plants through traditional two-dimensional PAGE and mass spectrometry identified <100 proteins (Kruft et al., 2001; Millar et al., 2001b). We have now expanded this identified set with direct sample analysis by liquid chromatography–tandem mass spectrometry (LC-MS/MS). This has allowed us to define a plant mitochondrial set of ∼400 proteins, of which nearly 20% are proteins of unknown function in Arabidopsis. We also compared our experimental set with the predictions of subcellular localization made by five targeting algorithms and with putative orthologs from other organisms. This analysis revealed new metabolic, regulation, and signaling pathways in plant mitochondria, providing a key data set for future experimental analysis of mitochondrial biogenesis, regulation, and function in plants.
RESULTS
MS Analysis of a Substantial Arabidopsis Mitochondrial Proteome
Previous evaluation of the whole Arabidopsis mitochondrial proteome using gel display and MS identified a nonredundant set of 92 proteins (Kruft et al., 2001; Millar et al., 2001b; Werhahn and Braun, 2002). A variety of more targeted studies of protein fractions and protein complexes partially purified from Arabidopsis mitochondria also have been undertaken (Sweetlove et al., 2002; Heazlewood et al., 2003a, 2003c; Herald et al., 2003; Millar and Heazlewood, 2003; Werhahn et al., 2003). Combining the data presented in all of these reports produces a nonredundant protein set of 145 experimentally determined Arabidopsis mitochondrial proteins. Our analysis of a membrane-enriched submitochondrial fraction from Arabidopsis cell culture using blue-native PAGE and MS/MS yielded many confirmatory identifications along with an additional 35 previously unidentified proteins (see supplemental data online). Altogether, 180 discrete proteins have been identified in Arabidopsis mitochondrial preparations using various gel-based arraying techniques.
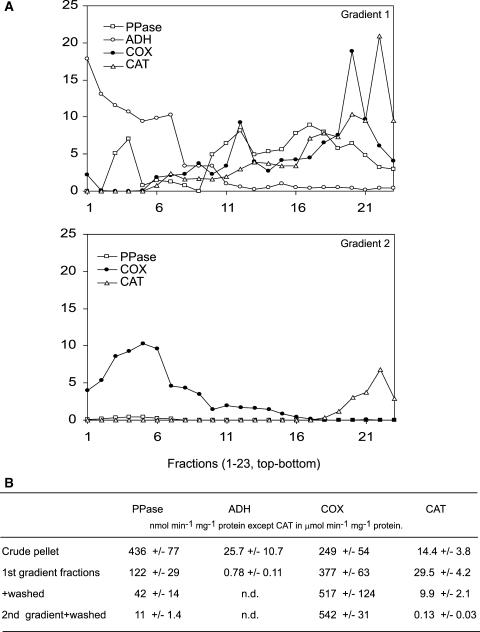
To overcome the clear limitations of these techniques, we undertook a systematic LC-MS/MS study of total Arabidopsis mitochondrial protein lysates. Mitochondria were isolated using two successive Percoll gradients designed to remove plastid and peroxisomal contaminants from crude organelle pellets derived from Arabidopsis cell cultures (Millar et al., 2001b) (Figure 1A). Using marker enzymes to determine the purity of these mitochondria, we showed that mitochondria were separated away from cytosolic, plastid, and peroxisomal contaminants (Figures 1A and 1B). Based on the calculations presented in Methods, the likely contamination on a protein basis in our purified mitochondrial samples was ∼1.5% for plastidic protein and less than ∼0.2% for peroxisomal protein (Figure 1B).
Figure 1.
Purification of Mitochondria from Arabidopsis Cell Cultures.
(A) Marker enzyme distribution in first and second density gradient separations of crude organelle pellets to yield purified mitochondria. Fractions 1 to 23 are 1.5-mL fractions from top to bottom of the gradients. Alkaline pyrophosphatase (PPase) is used as a plastid marker, cytochrome c oxidase (COX) is used as a mitochondrial marker, catalase (CAT) is used as a peroxisome marker, and alcohol dehydrogenase (ADH) is used as a cytosolic marker. Activities in each fraction are reported as percentages of activity found in crude organelle pellets.
(B) Specific activities of marker enzymes in the mitochondria-containing fractions at different stages of the purification process (means ± sd, n = 4). The crude organelle pellet protein is ∼60% plastidic, ∼30% mitochondrial, and ∼10% peroxisomal. Based on these proportions and the specific activities shown here, the final mitochondrial fractions contain ∼1.5% protein of plastidic origin and ∼0.2% protein of peroxisomal origin. n.d., not determined.
LC-MS/MS allows the direct identification of peptides derived from trypsin digestions of complex polypeptide samples, such as mitochondria, without the use of any preliminary gel separation of the polypeptides. This approach potentially can alleviate the problems of highly hydrophobic, basic, and small or very large molecular mass proteins being excluded from analysis, as often happens in polyacrylamide gel–based separations. Using high-stringency cutoffs for peptide matching in five independent mitochondrial samples isolated in this manner, the LC-MS/MS technique produced ∼2400 peptide MS/MS spectra that could be mapped redundantly to 390 Arabidopsis proteins.
An overall comparison of gel-based and LC-MS/MS methods indicated that >95% of the proteins identified previously by two-dimensional gel analysis of whole mitochondrial samples (Kruft et al., 2001; Millar et al., 2001b; Werhahn and Braun, 2002) were confirmed by LC-MS/MS (Table 1). Also, >80% of the proteins identified previously during purifications of particular subsets and protein complexes of mitochondrial proteins also were confirmed by our LC-MS/MS (Sweetlove et al., 2002; Heazlewood et al., 2003a, 2003c; Herald et al., 2003; Werhahn et al., 2003). Among 180 proteins defined by gel-based analysis, we confirmed 154 by LC-MS/MS (85%). The remaining 26 proteins identified previously using gel-based techniques were not found through the LC-MS/MS procedure (Table 1). A number of these 26 proteins were oxidative stress–induced proteins (Sweetlove et al., 2002); thus, their absence from our LC-MS/MS analyses of mitochondria from unstressed cells is understandable. This very high overlap between gel-array techniques and the LC-MS/MS method highlights the effectiveness of non-gel-based analysis and its broad application in proteomics. The data from the combination of these techniques yield a nonredundant Arabidopsis mitochondrial set of 416 proteins (154 defined by LC-MS/MS and gel separations, 26 by gel-based analysis only, and 236 by LC-MS/MS only; see supplemental data online). Of these, 407 are nucleus-encoded proteins and 9 are mitochondria-encoded proteins. Annotation of a relational database with these proteins allowed continued analysis of the set in the context of the whole Arabidopsis protein set. This database is now available publicly (http://www.mitoz.bcs.uwa.edu.au).
Table 1.
Functional Breakdown of the Arabidopsis Mitochondrial Set
| Function | Gels | Confirmed LC-MS/MS |
Novel LC-MS/MS |
Total LC-MS/MS |
Total |
|---|---|---|---|---|---|
| Energy | 73 | 66 | 25 | 91 | 98 (24%) |
| Metabolism | 37 | 33 | 44 | 77 | 81 (19%) |
| Protein fate | 20 | 19 | 33 | 52 | 53 (13%) |
| DNA synthesis and processing | 2 | 2 | 7 | 9 | 9 (2%) |
| Transcription | 1 | 0 | 7 | 7 | 8 (2%) |
| RNA processing | 3 | 3 | 11 | 14 | 14 (3%) |
| Protein synthesis | 4 | 3 | 11 | 14 | 15 (4%) |
| Defense, stress, detoxification | 8 | 6 | 8 | 14 | 16 (4%) |
| Communication/signaling | 3 | 1 | 16 | 17 | 19 (5%) |
| Transport | 13 | 12 | 6 | 18 | 19 (5%) |
| Miscellaneous | 2 | 2 | 2 | 4 | 4 (1%) |
| Structural organization | 1 | 0 | 8 | 8 | 9 (2%) |
| Unknown | 13 | 7 | 58 | 65 | 71 (17%) |
| Total | 180 | 154 | 236 | 390 | 416 |
The proportion of proteins in the 13 major functional groups are listed for the gel-based analysis of 180 proteins, the subset of these (154) confirmed by LC-MS/MS, the novel set found only by LC-MS/MS (236), and the total LC-MS/MS set (390). In total, this represents a set of 416 mitochondrial proteins (180 + 236).
Global Analysis of the Arabidopsis Mitochondrial Protein Set
Each member of the mitochondrial proteome set described above was assigned to a functional category using common functional divisions. Where annotations were unclear, a Basic Local Alignment Search Tool (BLAST) query against SWISS-PROT was undertaken for functional inference. This analysis produced 13 functional groups (Table 1). Known protein functions that are represented most highly in the mitochondrial proteome set are those involving energy (24%), metabolism (19%), and protein fate (13%). The proteins in these functional groups are generally well-characterized mitochondrial components of the tricarboxylic acid cycle, the respiratory chain, and the mitochondrial protein import, processing, assembly, and degradation machinery. The relative expression levels of proteins in these principal functional categories were profiled using EST numbers. More than 55% of the proteins that perform energy and metabolic roles and 30% of the proteins involved in protein fate are represented by >10 ESTs (data not shown). Nearly 75% of the energy group had been identified previously in gel-based separations, but the metabolism and protein fate groups contained many novel LC-MS/MS identifications. Approximately 25% of the identified proteins were involved in information-transfer processes (DNA/RNA synthesis and processing and protein synthesis), were defense/stress proteins, or were responsible for cellular interaction functions (transport, cell structure, and signal transduction proteins) (Table 1). The identification of proteins in these groups was aided by novel LC-MS/MS identifications. Only 34 proteins in these functional categories were identified in previous gel-based separations. We have confirmed 27 of these by LC-MS/MS and have added a novel set of 66 identifications to these sets of largely low-abundance components. Even when sequence similarity searches were undertaken against the nonredundant protein database at the National Center for Biotechnology Information, 71 proteins (17%) could not be confidently assigned a function and have been designated unclassified proteins. A similarly large set of unclassified proteins also was found in recent human and yeast mitochondrial proteome analyses (Sickmann et al., 2003; Taylor et al., 2003b).
The physical attributes of mitochondrial proteins identified solely by isoelectric focusing followed by SDS-PAGE often appear skewed compared with those of total proteomes because of the lack of identification of large and small proteins and the low detection rates of basic, hydrophobic, and low-abundance proteins (Heazlewood et al., 2003b). The use of LC-MS/MS greatly aided in the identification of large proteins (>80 kD) and proteins that were moderately hydrophobic (data not shown). The set of 416 mitochondrial proteins covered an isoelectric point range of 4 to 11, a molecular mass range of 6 to 428 kD, and a GRAVY (grand average of hydropathicity) range of −1.6 to +1.0 (−GRAVY = hydrophilic, +GRAVY = hydrophobic).
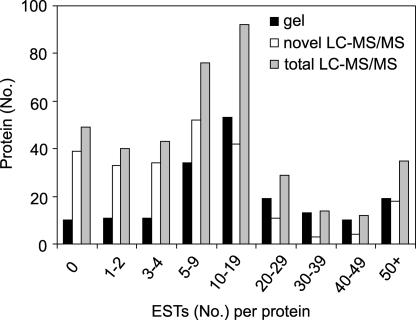
The representation of proteins in EST databases allowed an assessment of transcript abundance, and thus probable protein abundance, for different gene products. Most interestingly, there was a shift to much lower EST numbers for the new protein set identified only by LC-MS/MS (Figure 2). Nearly 45% of the novel identifications were represented by four or fewer ESTs (compared with only 17% of gel-based identifications). Some 39 newly identified proteins were so low in abundance that ESTs had not been reported by The Institute for Genomic Research (TIGR) at the time of writing (Figure 2). The large number of low-EST-number members in our protein set could be viewed as evidence that low-abundance paralogs, or even pseudogenes with high levels of identity to major protein isoforms, are clouding the analysis we have presented. A stringent analysis of all of the peptide sequences matched to proteins in our analysis was performed, and potential paralogs in our set are listed in the supplemental data online. These findings show that 15 proteins were identified that have no unique peptides that differentiate them from a highly similar paralog, whereas another 11 were identified with unique peptide sequences but the unique peptide(s) were of low matching confidence. There were also two cases in which all isoforms of some protein families were indistinguishable from each other as a result of very high sequence identity. However, the latter are well-characterized, abundant mitochondrial proteins—an elongation factor and the β-subunit of ATP synthase—that are tandemly arranged duplications on Arabidopsis chromosomes (see supplemental data online). All possible paralogs of this type are annotated in the supplemental data online. However, these possible paralogs represent only 16 of the 106 proteins identified with four or fewer ESTs and only 2 of the possible paralogs are among the 39 proteins with no registered ESTs. Thus, the low number of ESTs among the novel LC-MS/MS data broadly reflects low-abundance proteins within the mitochondrial proteome that originates from a low-abundance proteome within plant cells. This finding strongly suggests that a significant depth of the proteome has been revealed by our analysis: the identification of key low-abundance components. Many of these are in important areas of mitochondrial information transfer, regulation, and signaling.
Figure 2.
Low-Abundance Proteins Identified by LC-MS/MS.
Comparisons of the number of ESTs for each protein between the total gel-based proteomic analysis (180), the total LC-MS/MS set of proteins (390), and the novel proteins identified only by LC-MS/MS (236). EST numbers were derived from TIGR annotation data. All data are presented as numbers of nonredundant proteins in the EST number ranges indicated.
Specific Functions of the Arabidopsis Mitochondrial Proteome Set
The full set of experimentally identified proteins is provided in the supplemental data online or can be relationally searched directly at The Arabidopsis Mitochondrial Protein DataBase (http://www.mitoz.bcs.uwa.edu.au). We have identified many of the well-defined components of mitochondria, the electron transport chain, the tricarboxylic acid cycle, metabolic carriers, and the import apparatus. Among the remaining identifications, several prominent sets are worthy of more extensive comment, most notably the new identifications of low-abundance proteins with potentially key roles in mitochondrial function.
Cellular Signaling Components
There is increasing interest in intramitochondrial signaling pathways and also in the communication of mitochondria with the cytosol and other organelles, most notably the chloroplast and the nucleus. There also is extensive discussion of reactive oxygen species as signal molecules from mitochondria to other parts of the cell (Dutilleul et al., 2003) but little information on the pathways involved. Likewise, the presence of phosphorylated proteins in plant mitochondria has been documented (Bykova et al., 2003a, 2003b), but again, there is little information on the kinase/phosphatases involved or any associated signaling components that may regulate signaling through phosphorylation/dephosphorylation. Despite the importance of calcium in regulating mammalian mitochondrial function (Smaili et al., 2000), there is no direct evidence for such a role in plants. We have found mitochondrial protein components that may be involved in some of these processes. A set of 10 protein kinases, including Leu-rich repeat transmembrane protein kinases, receptor-like protein kinases, Ser-Thr kinases, and a mitogen-activated protein–like kinase, has been identified. No potential components of phosphorylation signaling pathways have been identified previously in plant mitochondria. Two of the Leu-rich repeat transmembrane receptor protein kinases (At1g74360 and At2g26330) show high sequence similarity to a protein kinase of unknown function found recently in human mitochondria (Taylor et al., 2003b). Nearly all of these kinases are very poorly represented in EST databases, all are individuals of large protein kinases subclasses, and none has a clear mitochondrial targeting sequence identified by bioinformatic predictors (Table 2). In addition to these discoveries is evidence for a 2A protein phosphatase regulatory subunit, a GTP binding protein, and several putative Ca2+ binding proteins. One of the putative Ca2+ binding proteins (At3g59820) is very similar in sequence to one found in mammalian mitochondria (Taylor et al., 2003b). A protein with sequence similarity to FRO1-like NADPH oxidase also was found. This protein generates superoxide from NADPH and has been linked to mitogenic signaling and cancer in smooth muscle cells in mammals (Suh et al., 1999).
Table 2.
Protein Kinases Identified in the Arabidopsis Mitochondrial Set
| AGI Locus No. | PPC Class | Description | PPC Subclass |
EST No. |
Targeting Prediction
|
||||
|---|---|---|---|---|---|---|---|---|---|
| Predotar | TargetP | MitoProt II | SubLoc | iPSORT | |||||
| Class 1: transmembrane receptor kinase and related nontransmembrane kinases | |||||||||
| At3g52530.1 | PPC:1.5.1 | Protein kinase-like protein | 23 | 1 | − | − | − | − | − |
| At4g23290.1 | PPC:1.7.2 | Ser/Thr kinase-like protein | 70 | 3 | C | S | − | − | M |
| At5g14210.1 | PPC:1.9.5 | Receptor protein kinase-like protein | 3 | 6 | − | S | − | − | Sig |
| At3g45420.1 | PPC:1.11.1 | Receptor-like protein kinase | 41 | 1 | − | S | − | S | Sig |
| At1g74360.1 | PPC:1.12.4 | Putative receptor protein kinase | 51 | 4 | − | S | − | − | − |
| At2g26330.1 | PPC:1.12.4 | Putative receptor-like protein kinase | 51 | 18 | − | S | − | − | Sig |
| At1g64210.1 | PPC:1.13.3 | Putative KRR receptor-like kinase | 14 | 1 | − | S | − | − | Sig |
| Class 2: ATN1/CTR1/EDR/GmPK6-like kinases | |||||||||
| At3g27560.1 | PPC:2.1.5 | Protein kinase ATN1 | 7 | 1 | − | − | − | M | − |
| Class 4: nontransmembrane protein kinases | |||||||||
| At1g69220.1 | PPC:4.1.2 | Putative STE20/PAK-like protein kinase | 11 | 3 | − | − | − | N | − |
| At3g18040.1 | PPC:4.5.1 | Putative mitogen-activated protein kinase | 23 | 2 | − | − | − | N | − |
Descriptions of the 10 protein kinases identified are shown in their three different PlantsP Kinase Classification (PPC; http://plantsp.sdsc.edu/plantsp/family/class.html) classes. The total number of gene members in each subclass is shown, along with the specific AGI number of the mitochondrial member identified. The number of ESTs for the identified kinases and their predicted subcellular locations by the five targeting prediction programs are shown. C, chloroplast; M, mitochondria; N, nucleus; S, secretory pathway; Sig, signal sequence; −, unknown (no target or cytosolic).
Regulation of Protein Complex Assembly
More than 50 proteins involved in protein fate were identified. These proteins spanned protein import and sorting, presequence cleavage, molecular chaperone, complex assembly, and proteolysis functions. Protein components of the LON, Clp, and FtsH protease systems and three other putative proteases were identified. Seven subunits from the translocases of the outer and inner membranes (TOM and TIM) of the import apparatus also were found. Interestingly, four proteins with sequence similarity to components implicated specifically in the assembly of respiratory electron transport chain complexes were found. These included At5g51220 and At1g07510/At3g17910, which are similar to two yeast factors required for the assembly of complex III of the respiratory chain (C3BP and BCS1, respectively). We also found a putative cytochrome c oxidase assembly factor similar to the yeast SHY1/surf1 protein. The absence of SHY1 in yeast leads to <30% cytochrome c oxidase assembly, while mutations of the human ortholog (surf1) lead to Leigh syndrome (Nijtmans et al., 2001). Cytochrome c biogenesis in plant mitochondria occurs by the system-I mechanism that is shared in common with bacteria but differs from the systems used in chloroplasts (system II) and fungal, vertebrate, and invertebrate animal mitochondria (system III) (Kranz et al., 1998). Although some of the system-I components are encoded in the mitochondrial genomes of plants, others have been transferred to the nucleus. We have identified a protein similar to the bacterial cycL/ccl2/ccmH subunit, which is critical for conferring specificity to the thio-redox pathway of cytochrome c biogenesis. To our knowledge, this plant mitochondrial CCMH-like gene (At1g15220) has not been described in plant studies to date. We also found two proteins similar to the yeast AFG3 metalloprotease that is of the AAA-type ATPase family. These proteases have been implicated in the regulation of the electron transport chain through the degradation of unincorporated or incompletely synthesized mitochondria-encoded proteins in yeast (Guzelin et al., 1996).
Key Proteins Involved in DNA Synthesis, Transcription, and RNA Binding/Processing
Low-abundance proteins from these classes have been poorly represented in proteomic studies of plant mitochondria to date. A DNA polymerase and several DNA gyrase subunits were localized to mitochondria in this study. Several differentiation and greening (DAG)-like proteins also were found. Although DAG proteins were associated initially with chloroplasts, a member of this gene family has been reported in two-dimensional PAGE analyses of plant mitochondria (Kruft et al., 2001). In both snapdragon and Arabidopsis, the absence of specific DAG proteins greatly influences chloroplast differentiation and palisade tissue development (Chatterjee et al., 1996; Babiychuk et al., 1997; Bisanz et al., 2003). However, to our knowledge, the functions of these proteins in mitochondria have not been reported. Transcription initiator and terminator factors, and a series of proteins from transcription factor gene families, also were identified. We also found a protein (At5g12290) highly similar to a protein responsible for the regulation of the abundance of transcripts from mitochondrial respiratory genes in yeast. NCA2 is a nucleus-encoded protein involved in the turnover of ATP6/ATP8 transcripts in yeast mitochondria (Camougrand et al., 1995). We also identified peptides from several RNA helicases and 10 pentatricopeptide repeat proteins. The latter are highly represented in plant genomes and have roles as restorers of cytoplasmic male sterility (Bentolila et al., 2002) and putative roles in RNA editing (Small and Peeters, 2000).
Carriers Essential for Whole Plant Growth and Bypasses of the Classic Electron Transport Chain
We have identified several proteins involved in iron metabolism, including two putative ABC transporters (At4g28620 and At5g58270) with strong sequence similarity to yeast ATM1, the mitochondrial Fe-S transporter (Kispal et al., 1997). We also identified a Cys desulfurase (At5g65720) with sequence similarity to Nfs1 from yeast (the yeast ortholog of bacterial NifS). In yeast, these proteins act in concert for the synthesis and transport of Fe/S clusters and have been shown to play an essential role in the synthesis of both intramitochondrial and extramitochondrial Fe/S proteins (Kispal et al., 1999). In plants, forward genetics identified this ABC transporter gene (At5g58270) as the basis for the starik mutant, which has a dwarfism/chlorosis phenotype (Kushnir et al., 2001). We also identified eight carriers of the mitochondrial carrier superfamily, including two additional carriers not detected in a previous analysis of this superfamily (Millar and Heazlewood, 2003). One of these new identifications has strong similarity to carnitine-acylcarnitine carriers (At5g46800), and the other is a mitochondrial carrier of unknown function (At5g15640). In mammalian mitochondria, carnitine acyl carrier proteins transport lipid-derived molecules across mitochondrial membranes for energy and carbon supply. The carnitine-acylcarnitine carrier was revealed recently as the gene responsible for the à bout de souffle Arabidopsis mutation (Lawand et al., 2002). This mutant plant stops developing after germination, degrades storage lipids, and develops further only when germinated in the dark or after the addition of exogenous sugar (Lawand et al., 2002). We have direct evidence for the presence of enzymes putatively responsible for the bypasses of the respiratory chain in plants, with proteins for rotenone-insensitive NAD(P)H-dependent ubiquinone oxidoreductases and the alternative oxidase reported. We also show that both the α- and β-subunits of the electron transport flavoprotein (At1g50940 and At5g43430) are present in Arabidopsis mitochondria. These proteins are responsible for direct electron transfer from fatty acid to ubiquinone and branched chain amino acid oxidation in mammals (Frerman, 1987), but they have not been investigated in plants.
Proteins of Unknown Function
In addition to the proteins described above, 71 proteins were identified for which no clear function could be assigned. Only 13 of these proteins were identified in previous gel-based proteome analyses (Table 1). The identification of these proteins provides a basis for the use of reverse genetics to identify novel mitochondrial functions in plants.
Comparing Mitochondrial with Plastid and Peroxisomal Proteomes
To determine whether the identified proteins are truly mitochondrial or to what degree some proteins are from low-level contamination in our mitochondrial samples from other cellular compartments, we compared our analysis to those performed for other organelles from Arabidopsis (see supplemental data online). Three catalase proteins have been identified in our mitochondrial set, and although several claims of low levels of mitochondrially localized catalase in plants have been made, these proteins typically represent the bulk of peroxisomal protein. Two catalase proteins (At1g20620 and At4g35090) in our mitochondrial set are among the set of highly abundant proteins identified by Fukao et al. (2002) in their peroxisomal proteome analysis. None of the other peroxisomal proteins noted by Fukao et al. (2002) were found in our mitochondrial analysis. Of the 461 nonredundant proteins identified definitively in large-scale thylakoid, lumen, and envelope preparations from Arabidopsis by MS (Peltier et al., 2002; Schubert et al., 2002; Ferro et al., 2003; Froehlich et al., 2003), only 31 are found in our mitochondrial set. These are presented in the supplemental data online, where they are dissected into probable mitochondrial contaminants in chloroplast preparations (12), probable chloroplast contaminants in mitochondrial preparations (12), unknown location (4), and probable peroxisomal contaminants in mitochondria and chloroplasts (3). Thus, there are 15 proteins that probably represent a combination of chloroplast and peroxisomal contamination. These published sets from other organelles also are available for relational searches in a database we have established (http://www.mitoz.bcs.uwa.edu.au). There is the further possibility that some of these apparent contaminants could be dual-targeted proteins in plants, reflecting their presence in multiple compartments (Peeters and Small, 2001). We recently showed that the stromal ascorbate peroxidase and stromal monodehydroascorbate reductase identified in this LC-MS/MS experiment (At4g08390 and At1g63940) are authentically dual targeted to chloroplasts and mitochondria in Arabidopsis (Chew et al., 2003). We also have shown that a range of glycolytic enzymes identified here by LC-MS/MS are present in our samples, because glycolytic enzymes found in the cytosol also are associated functionally with the outer membrane of plant mitochondria (Giegé et al., 2003).
Orthologous Comparisons of Mitochondrial Proteomes
Because the endosymbiosis of the mitochondrial progenitor by the ancestral eukaryotic cell occurred before the divergence of plants and animals (Gray et al., 2001), cross-kingdom similarity comparisons can be used to identify putative orthologous proteins with conserved function. Mitochondrial protein sets from both yeast (552 proteins) (Schon, 2001) and human (615 proteins) (Taylor et al., 2003b) are available, as is the protein set from the closest living relative to the mitochondrial progenitor, the bacterial endosymbiont Rickettsia prowazekii (834 proteins) (Andersson et al., 1998). These three sets were compared with the entire Arabidopsis proteome using the program INPARANOID (Remm et al., 2001). This program searches for high-stringency orthologous clusters between two protein sets, providing clusters of paralogs within species and orthologs across species. Gene matching using this procedure is considered to exclude many lower similarity matches for which true cluster orthology, and thus conserved functionality, is questionable.
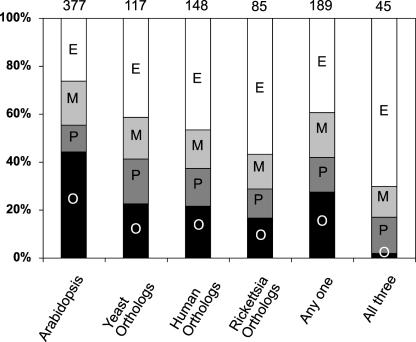
A reduced mitochondrial set of 377 proteins that excluded the 26 possible paralogs (see supplemental data online) and the partially overlapping set of 15 probable contaminants (see supplemental data online) was constructed and used for this orthology comparison. A total of 117 yeast mitochondrial proteins were identified as putative orthologs of this reduced Arabidopsis mitochondrial protein set. More than 75% of the putative orthologs fall into the major functional groups of energy, metabolism, and protein fate, which encompass only 55% of the total Arabidopsis mitochondrial set (Figure 3). Similar comparisons with the human mitochondrial proteome and the Rickettsia protein set identified 148 putative orthologs to human mitochondrial proteins and 85 putative orthologs to Rickettsia proteins within the Arabidopsis mitochondrial set. In total, 50% (189 proteins) of the Arabidopsis mitochondrial set had putative orthologous matches with at least one protein from yeast mitochondrial, human mitochondrial, or Rickettsia proteomic sets (Figure 3, any one). The putative orthology networks derived from these comparisons has a strict common overlap of 45 proteins (Figure 3, all three), in which an individual Arabidopsis protein has a putative ortholog in the protein set from each of the three other species. This forms a central cluster of putative orthologs between the Rickettsia set and the current mitochondrial proteomes of divergent organisms. This common set is very heavily skewed toward the functional category of energy (70%), with several members from the metabolism (13%) and protein fate (15%) categories (Figure 3).
Figure 3.
Major Functional Distributions of the Arabidopsis Mitochondrial Set and of Matching Putative Orthologs from Yeast, Human, and Rickettsia.
Orthologs were determined through high-stringency sequence homology matching using the program INPARANOID (Remm et al., 2001). The distribution of putative orthologs to the reduced Arabidopsis mitochondrial set of 377 proteins is presented for the three major functional classes of energy (E; white), metabolism (M; light gray), and protein fate (P; dark gray) and for all other identifications (O; black). The total number of putative orthologs in each species, in at least one set, and in all three sets is shown above each bar.
We found very different putative orthology patterns when we considered the low-abundance proteins involved in mitochondria–cellular interaction (signaling, transport, and structure) and information-transfer processes (DNA synthesis and processing, transcription, and RNA binding/processing) (Table 3). Although putative orthologous proteins were present in most of these functional classes, the overlap between different organisms was very low, and no one protein common to each of the protein sets was found by INPARANOID analysis. The kinases observed in human and plant mitochondria were absent from yeast mitochondrial sets, whereas plant transporters and transcriptional regulators present in the yeast set were not in the human set. Interestingly, the DNA gyrase subunits A and B found in Arabidopsis mitochondria have putative orthologs among Rickettsia proteins but not among yeast or human mitochondrial proteins. Significantly, the large set of Arabidopsis proteins of unknown function do not have many putative orthologs in other species, suggesting that they are likely to be largely plant-specific mitochondrial proteins (Table 3).
Table 3.
Minor Functional Group Distributions of the Arabidopsis Mitochondrial Set with Matching Sequence Orthologs from Yeast, Human, and Rickettsia
| Species | Communication and Signaling |
Transport | Cell Structure |
DNA Synthesis and Processing |
Transcription | RNA Binding/Processing |
Unknown Function |
|---|---|---|---|---|---|---|---|
| Arabidopsis | 17 | 17 | 6 | 8 | 8 | 14 | 69 |
| Yeast orthologs | 0 | 10 | 1 | 0 | 1 | 2 | 0 |
| Human orthologs | 4 | 10 | 2 | 0 | 0 | 0 | 4 |
| Rickettsia orthologs | 1 | 2 | 0 | 2 | 0 | 0 | 2 |
| Any one | 4 | 13 | 2 | 2 | 1 | 2 | 5 |
| All three | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
Orthologs were determined through high-stringency sequence homology matching using the program INPARANOID (Remm et al., 2001). The 139 proteins in the stringent mitochondrial set of 377 proteins in the functional classes of communication, transport, cell structure, DNA synthesis and processing, transcription, RNA binding/processing, and unknown function are represented along with putative ortholog numbers in different species. The total number of Arabidopsis proteins in each set with any ortholog is shown (any one) as well as the total number with orthologs in all three other species (all three).
Comparison of Experimental and Bioinformatic Predictions of the Arabidopsis Mitochondrial Proteome
We compared our reduced experimental set of 377 proteins, which contained 368 nucleus-encoded proteins, with those proteins predicted to be mitochondrial by publicly available subcellular targeting prediction programs: TargetP, Predotar, MitoProt II, SubLoc, and iPSORT. The number of nucleus-encoded Arabidopsis proteins predicted by each program to be targeted to mitochondria ranged from 3000 to 4500 (Table 4). The ability of the various targeting programs to correctly predict the experimental Arabidopsis mitochondrial set was significantly less than expected (Table 4). The successful prediction rates are presented as sensitivity, defined as the percentage of the experimental set predicted correctly (Table 4). Sensitivities of these programs were in the range of 37 to 47% for the set of 368 proteins. We then defined an even more reduced set of proteins that have been found in gel-based proteome analysis and also in our LC-MS/MS experiments: these are 143 higher abundance proteins with very high identification confidence values. In this set, the sensitivities of the programs increased to 49 to 68%. For comparison, we also considered the prediction sensitivities of TargetP and MitoProt II on the defined mammalian and yeast mitochondrial sets (Schon, 2001; Taylor et al., 2003b). These showed sensitivities of 42 to 59% depending on the predictor and the species (data not shown). Another recent analysis of the yeast experimental mitochondrial proteome set with MitoProt II showed that only 43% of proteins contain predicted N-terminal presequences (Sickmann et al., 2003). All of these values are significantly lower than the previously reported sensitivities of 80 to 90% for mitochondrial prediction that generally are attributed to these programs (Emanuelsson et al., 2000; Hua and Sun, 2001; Bannai et al., 2002).
Table 4.
Evaluation of Mitochondrial Prediction in Arabidopsis by Bioinformatic Subcellular Predictors
| Program | Total Predicted | Mitochondrial Set (368) | Sensitivity | Mitochondrial G/LC Set (143) | Sensitivity |
|---|---|---|---|---|---|
| Predotar | 4514 | 169 | 46% | 95 | 66% |
| TargetP | 2955 | 145 | 39% | 88 | 62% |
| MitoProt II | 3794 | 173 | 47% | 97 | 68% |
| SubLoc | 3554 | 135 | 37% | 70 | 49% |
| iPSORT | 4492 | 174 | 47% | 94 | 66% |
Performance comparison of the five subcellular predictors with the Arabidopsis experimentally derived mitochondrial set. Two mitochondrial sets were used in the analysis: a set of the 368 nucleus-encoded mitochondrial proteins after the removal of paralogs and potential nonmitochondrial contaminants (see supplemental data online), and a subset of 143 nucleus-encoded mitochondrial proteins from the set of 368 that were identified in both LC-MS/MS and gel experiments. Sensitivity is the percentage of this set predicted by each program. This analysis excluded the nine mitochondria-encoded proteins identified in our analysis.
The use of only well-known mitochondrial proteins in the initial neural network training of these programs may have contributed to the previously reported high percentages and the increases observed when only the major, well-known proteins of mitochondria were considered in our analysis (Table 4). In the case of the rule-based MitoProt II, sensitivity was assessed previously using a set of proteins that also would have been examined in the initial appraisal of the rules for organellar targeting (Claros and Vincens, 1996). That is, the previous values are based on a certain degree of circular logic and rely on a known set of largely matrix-targeted mitochondrial proteins with classic N-terminal presequences that does not truly represent the breadth of the real mitochondrial proteomes in plants, animals, or fungi. Our experimental data set includes noncleaved proteins and/or those with internal, C-terminal, or cryptic targeting signals that have not been incorporated in previous test sets for sensitivity. Sensitivity, however, also must be balanced by the size of the total protein set from the genome predicted by a program to be mitochondrial (Table 4). Based on such comparisons, the program TargetP predicts the largest positive set from the smallest total pool, whereas the program Predotar is least efficient at predicting a similar number of our experimental set, but from a larger total pool. The program with the best combination of sensitivity and total prediction set size for mitochondria in Arabidopsis appears to be MitoProt II, whereas the worst performing program was SubLoc. A new version of Predotar (version 1.03) recently released on the World Wide Web (http://genoplante-info.infobiogen.fr/predotar) provides a much smaller set size in Arabidopsis (1148) but predicts only 121 of the proteins in our experimental set (sensitivity of 33%).
To further assess prediction program performance, we assessed the overlapping prediction of the Arabidopsis protein set by the five predictors. In recent studies of mitochondria, combinations between TargetP, Predotar, and MitoProt II have been selected empirically as useful sets for fast tracking the identification of possible mitochondrial functions in plants that then require experimental investigation (Kruft et al., 2001; Millar et al., 2001b; Elo et al., 2003; Heazlewood et al., 2003b). Using relational searches, a comparison of these programs predicting mitochondrial localization was undertaken (Table 5). The number of proteins uniquely predicted as mitochondrial by each of the five programs ranged from 400 to 2000 depending on the predictor (Table 5, positive column). The program SubLoc shared the least similarity in its targeting prediction to the other four programs. SubLoc predicted 1999 proteins, representing >75% of its total prediction set, that are not predicted to be mitochondrial by any other program. By contrast, the set of mitochondrial targeted proteins predicted by TargetP had the highest shared predictive set with the other programs, with only 424 proteins (14% of the total TargetP set) predicted by this program alone. To find consensus, an analysis was undertaken of the proteins uniquely predicted not to be localized to the mitochondria by a given program while the other four programs predicted a mitochondrial localization (Table 5, negative column). Only SubLoc displayed a significant lack of accord in its prediction pool, with 638 proteins uniquely predicted not to be mitochondrial, whereas the other four programs were in agreement.
Table 5.
Prediction of Mitochondrial Localization in Arabidopsis
| Program | Total | Percentage | Positive | Negative |
|---|---|---|---|---|
| Predotar | 4514 | 16.5 | 1371 | 61 |
| TargetP | 2955 | 10.8 | 424 | 63 |
| MitoProt II | 3794 | 13.9 | 942 | 65 |
| SubLoc | 3554 | 13.0 | 1999 | 638 |
| iPSORT | 4492 | 16.5 | 1266 | 58 |
An analysis of mitochondrial localization was undertaken using the entire Arabidopsis protein set consisting of 27,288 nucleus-encoded proteins with five subcellular prediction programs. Total indicates the total number of proteins each predictor assigns as localized to the mitochondria and its relative percentage of the total proteome. Positive prediction numbers indicate the number of proteins predicted as mitochondrial only by this program compared with the others. Negative prediction numbers indicate the number of proteins predicted as mitochondrial by all other programs but not this program.
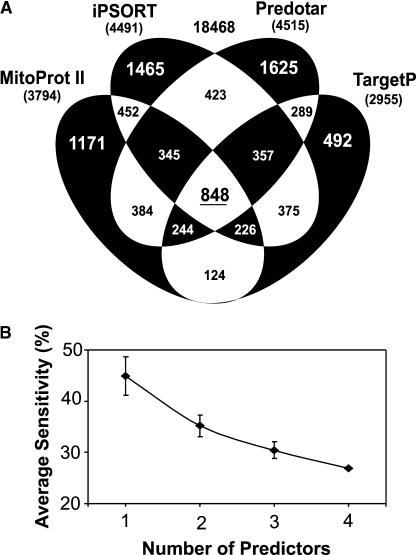
When the overlap of prediction between any two programs was assessed, it could be as little as 20% (TargetP/SubLoc) and averaged only ∼45% (Tables 5 and 6). Overlap of more than two program prediction sets caused further reductions, to yield sets containing only 1 to 4% of the Arabidopsis protein set. The exclusive overlapping sets of predicted proteins are shown graphically in Figure 4A as a four-way Venn diagram highlighting the large sets of unique predictions by each program and the proportion of each exclusive intersect between prediction programs (the program SubLoc was excluded from this analysis because of its poor performance; see above). This Venn diagram analysis allowed us to determine that 8820 proteins, representing 32% of the total predicted protein set from Arabidopsis, are predicted to contain a mitochondrial localization signal recognized by at least one of the four better prediction programs. A mitochondrial proteome of this size is inconsistent with estimates of 5 to 10% of nucleus-encoded protein sets (typically 500 to 2000 proteins) proposed in other eukaryotes and would not be given credence by researchers. On the other hand, all four predictors agree on 848 members of this protein set (Figure 4A). This analysis shows that assigning probable mitochondrial localization based on only one or two prediction programs is unlikely to be accurate, because the choice of program can greatly influence the pool of proteins selected. The accuracy of prediction is likely to be highest in the set common to all of the prediction programs.
Table 6.
Relational Combinations of the Five Prediction Programs Indicating the Number and Proteomic Percentage of Shared Proteins That They Predict To Be Localized to the Mitochondria
| Program Combinations | Total | Percentage |
|---|---|---|
| MitoProt II/iPSORT | 1872 | 6.9 |
| Predotar/MitoProt II | 1820 | 6.7 |
| Predotar/iPSORT | 1974 | 7.2 |
| Predotar/TargetP | 1738 | 6.4 |
| TargetP/iPSORT | 1806 | 6.6 |
| TargetP/MitoProt II | 1442 | 5.3 |
| MitoProt II/SubLoc | 802 | 2.9 |
| SubLoc/iPSORT | 800 | 2.9 |
| Predotar/SubLoc | 842 | 3.1 |
| TargetP/SubLoc | 583 | 2.1 |
| Predotar/MitoProt II/iPSORT | 1194 | 4.4 |
| TargetP/Predotar/iPSORT | 1205 | 4.4 |
| Predotar/MitoProt II/TargetP | 1092 | 4.0 |
| MitoProt II/TargetP/iPSORT | 1074 | 3.9 |
| Predotar/MitoProt II/SubLoc | 404 | 1.5 |
| iPSORT/MitoProt II/SubLoc | 421 | 1.5 |
| iPSORT/Predotar/SubLoc | 404 | 1.5 |
| TargetP/MitoProt II/SubLoc | 349 | 1.3 |
| Predotar/TargetP/SubLoc | 386 | 1.4 |
| iPSORT/TargetP/SubLoc | 384 | 1.4 |
| Predotar/MitoProt II/TargetP/iPSORT | 848 | 3.1 |
| iPSORT/SubLoc/TargetP/Predotar | 275 | 1.0 |
| Predotar/MitoProt II/TargetP/SubLoc | 268 | 1.0 |
| TargetP/MitoProt II/SubLoc/iPSORT | 271 | 1.0 |
| Predotar/MitoProt II/SubLoc/iPSORT | 273 | 1.0 |
| Predotar/MitoProt II/TargetP/iPSORT/SubLoc | 210 | 0.8 |
Figure 4.
Evaluation of Mitochondrial Prediction in Arabidopsis by Intersections between Targeting Prediction Programs.
(A) Venn diagram illustrating the overlap of mitochondrial prediction of the Arabidopsis nucleus-encoded proteome comprising 27,288 proteins (TIGR ATH1 release 3.0 including supplementary data). The subcellular prediction programs MitoProt II, iPSORT, Predotar, and TargetP were chosen for whole-proteome analysis because they all displayed similar sensitivities with regard to mitochondrial localization (Table 2). The number of mitochondria-targeted proteins predicted by all four programs is underlined (white central section); the six other white areas indicate two-way predictive relationships between the programs. The inner black regions indicate three-way predictions, and the outer black areas indicate the number of novel proteins predicted by each program. Numbers in parentheses represent the total number of mitochondrial proteins predicted by each program. The number outside the Venn diagram (18,468) represents the null set.
(B) Relational interaction between averaged sensitivities using multiple predictors with the experimentally derived nonredundant set of 368 nucleus-encoded proteins from Arabidopsis. The use of multiple predictions in the assessment of mitochondrial localization results in a substantial reduction in sensitivity. Standard deviations are presented as error bars: one predictor, n = 4; two predictors, n = 6; three predictors, n = 4; four predictors, n = 1.
We also used relational searches to compare directly the overlapping mitochondrial prediction sets from bioinformatic tools with our reduced experimental data set of 368 nucleus-encoded proteins in an attempt to determine the most accurate overlapping combination of predictor programs for mitochondria. We found that although the total set predicted decreased when multiple predictors were used together (Figure 4A), the value of this was offset substantially by degradations in sensitivity (Figure 4B). Consequently, the use of multiple relational predictions generates an enriched mitochondrial pool but also excludes many genuine mitochondrial proteins that we observed in the experimental set. Thus, if the sequence of an unknown protein is predicted to be mitochondrial by a number of different programs, the probability that it is mitochondrial increases; however, the lack of prediction by any or by only some programs by no means precludes a mitochondrial destination.
DISCUSSION
We present a large-scale proteomic and bioinformatic analysis of the Arabidopsis mitochondrial proteome. This data set allows us to consider some very fundamental biological issues pertaining to mitochondria in plants that have a significant impact on how these organelles are viewed and how future research on them can be facilitated. First, it suggests a novel selection of genes that regulate and coordinate mitochondrial biogenesis and function in plants. Second, the unique features of plant mitochondria that set them apart from their counterparts in other organisms can be revealed systematically. Third, the veracity of bioinformatic targeting predictors can be assessed directly, allowing decisions to be made about whether it is necessary to more extensively assess subcellular proteomes experimentally in plants or whether targeting prediction from protein sequences is sufficient.
Identifying Key Mitochondrial Targets for Genetic Manipulation
Reverse genetics approaches are likely to be very powerful tools in the study of mitochondrial function in plants. Importantly, these approaches require gene-specific identification of candidates for genetic manipulation. Subcellular proteomics provides a valuable means of providing this gene-specific information, cutting through the complexity of gene families and supergene families to direct the strategies for genetic manipulation. Subcellular proteomics does this by providing a protein snapshot of where and often in what abundance specific proteins accumulate in different cellular compartments. Initial attempts to experimentally derive the genes that defined plant mitochondrial proteomes in plants using gel-based techniques resulted in the identification of only ∼100 to 150 proteins from several different species (Kruft et al., 2001; Millar et al., 2001b; Bardel et al., 2002; Werhahn and Braun, 2002; Heazlewood et al., 2003b). The well-known relatively abundant enzymes of mitochondrial metabolism predominated in these analyses, and few truly new insights into mitochondrial function were obtained. The LC-MS/MS analysis presented here provides the next level of proteome resolution. It looks below the metabolic enzymes to reveal transcriptional regulators, protein complex assembly factors, low-abundance carriers, minor-flux biosynthetic pathways, and potential signaling components. These components often are present in gene families of significant size in plants; thus, the detection of specific gene family members provides a very significant advance and direct targets for genetic manipulation, functional verification, and mutant phenotype assessment. For example, the 10 protein kinases identified here are members of complex multigene families that contain 243 members (Table 2) and do not contain recognizable targeting presequences. Proteomics was the only reasonable means to readily identify this mitochondrial protein kinase complement.
Origins, Divergence, and Specialization of the Mitochondrial Proteome
It has been suggested that a large proportion of mitochondrial proteins evolved from prokaryotic lineages (50 to 60%), with the remaining proteins constituting a eukaryotic subset (20 to 30%) and a speculative species-specific subset (20%) (Karlberg et al., 2000; Marcotte et al., 2000). These studies were undertaken using similarity-based techniques, and as a result, the overlap between these groups is likely to be somewhat less distinct in a strictly evolutionary sense (Gray et al., 2001). Nonetheless, a similarity approach provides a valuable framework for considering how the proteomes of mitochondria and related microbes differ and thus provides a glimpse into the history of the development of subcellular proteomes. We used an approach that specifically seeks to identify orthologs in other species (Remm et al., 2001). A comparison of the Arabidopsis mitochondrial protein set with the human and yeast mitochondrial protein sets and the proteome of Rickettsia, the nearest living relative of the mitochondrial progenitor, indicated that they share a substantial set of putative orthologs in other species (Figure 3). Fifty percent of the Arabidopsis mitochondrial protein set can be linked through putative orthology to sequences in other mitochondria or free-living mitochondria-like organisms. This is a stringent set of identifications that excludes a significant number of proteins predicted to have similar functions on the basis of simple sequence similarity comparisons, especially those based on the presence of sequence-derived functional motifs between different species.
Within these putative orthology sets, the major metabolic groups of energy, metabolism, and protein fate predominate (Figure 3). The group shared between eukaryotic mitochondrial and Rickettsia protein sets displays this bias even more prominently (Figure 3). Thus, the central bioenergetic and metabolic function of mitochondria appears to be evolutionarily more highly conserved, although genes that encode proteins involved in other functions may have diverged more widely in sequence and/or may have been more readily exchanged with proteins encoded in host (nucleus) and coendosymbiont genomes. This divergent set with low orthology rates was very clear in the analysis of mitochondrial–cellular interaction proteins (signaling, transport, and structure) and proteins of information-transfer processes (DNA synthesis and processing, transcription, and RNA binding/processing). Thus, although the central functions of mitochondria are conserved, the regulation of the mitochondrial genome, cellular signaling, and transport between the mitochondrion and the cell have diverged. Consequently, plant mitochondria appear to be integrated in the function of the whole cell in a way that uses some proteins in common with mitochondria from other organisms but that also has involved recruiting novel proteins to play roles in mitochondria that are unique to plants.
The 20% of the yeast mitochondrial proteome that is thought to be unique to this species (Marcotte et al., 2000) is similar to the proportion of proteins of unknown function in the Arabidopsis and human mitochondrial sets. There is little putative orthology overlap between these sets of proteins with unknown function (Table 3), and it is possible that they reflect mitochondrial functions that are unique to each of the three species. The likelihood of large species-specific protein sets also was highlighted in the recent comparative genomic analysis of mitochondrial proteomes by Richly et al. (2003). In our view, functional investigation of this unknown-function set and the set of putative-function proteins that lack putative orthologs outside of plants will provide the key to enhancing our understanding of mitochondria from a uniquely plant perspective.
The Veracity of Prediction Programs for Mitochondrial Protein Localization
Computer-based prediction programs are used widely to predict a protein's function and/or location within the cell. In conjunction with the experimental and orthology-based methods discussed above, we have attempted to profile a theoretical plant mitochondrial proteome using the publicly available cell localization prediction programs TargetP, Predotar, MitoProt II, SubLoc, and iPSORT. The finding that approximately one-third of the entire Arabidopsis protein set is predicted to be localized to the mitochondria by any one predictor came as a surprise and suggests that these programs predict mitochondrial targeting rather loosely. The well-established programs TargetP, Predotar, and MitoProt II displayed ∼60% overlap with each other and with the more recently developed program iPSORT. By contrast, SubLoc predicted subsets that share only ∼40% identity with the other programs. Overall, these predictors identified ∼40 to 70% of the experimental mitochondrial proteome (Table 4). In our view, researchers should be cautious in the inferences drawn and the credibility given to prediction sets derived from these programs. The development of new or improved predictors seems necessary, and large experimental sets like the one presented here will form a valuable basis for training and assessing neural networks. An attractive alternative, at least for model plants, will be an alliance of large-scale fluorescence tagging, epitope tagging, and ever more subfractionation of subcellular proteomics to provide experimental evidence on a genome-wide scale, providing more clear-cut information for researchers.
Integration of Proteomic Data with Genomic Resources in Arabidopsis
With the increased use of proteomics and the enormous amounts of data being produced through genomic and microarray experiments, there is an urgent need for the development of new methods for the analysis and dissemination of these data in a genomic context. There currently exist several Arabidopsis genomics-based databases that are concerned with primary sequence annotation, namely TIGR, MIPS, and The Arabidopsis Information Resource (TAIR). Recently, TAIR has begun to develop queries that allow for the interrogation of the TIGR Arabidopsis database for a variety of basic protein attributes. The incorporation of biochemical pathways (AraCyc), protein motifs, and gene ontologies in the current Arabidopsis annotation release has expanded the use of TAIR for interactive protein queries. Nonetheless, one of the underlying problems that emerge when any of these databases is interrogated is the confusion caused by the growing families of proteins without evidence of protein functionality and without the context of cellular compartmentalization. Experimental proteomics has been severely limited to date by how data have been archived, annotated, manipulated, and presented. Currently, most data are available as text lists of identified proteins, and at best these lists are linked to “two-dimensional gel reference maps.” Attempts are under way by the wider proteomic community to address some of these issues (Taylor et al., 2003a), although these ideas are likely to take some time to be adopted on a large scale and across many organisms. We have developed a relational database for the analysis and presentation of experimental proteomic data alongside genomic data and bioinformatic predictions. The inclusion of the entire Arabidopsis predicted proteome in our database also enables this resource, or clones of it, to be expanded further to cover global Arabidopsis proteomics and/or other subcellular proteomes. We also incorporated the published data on chloroplast and peroxisomal proteomes (Fukao et al., 2002; Peltier et al., 2002; Schubert et al., 2002; Ferro et al., 2003; Froehlich et al., 2003) for additional relational searching.
Future Proteomic, Transcriptomic, and Genomic Profiling of Plant Mitochondria
Finally, it should be noted that the plant mitochondrial proteome is not a static entity. The overall proteome is a dynamic set of proteins that change over time between different cells and tissues. Nonetheless, it is possible to identity a set of proteins that is found in the mitochondria at a given time in a particular tissue. The Arabidopsis mitochondrial proteome subset described in this study is derived from cultured cells, but it is likely to contain a substantial number of the proteins that eventually will be found to constitute the entire Arabidopsis mitochondrial proteome. This set contains proteins identified from a multitude of other studies to be associated definitively with the mitochondria but also contains many previously unidentified proteins of unknown function, including an array of important low-abundance signaling and regulatory components. The mitochondrial proteome set will be expanded online as additional experimental evidence is gathered and collated. Microarray data generated under various conditions that influence mitochondrial function will be incorporated into this database in the future to allow dynamic searching and clustering of the mitochondrial proteome with changes in transcript abundance. This will allow the definition of common patterns of expression and thus putative regulators of mitochondrial biogenesis, stress response, and other aspects of nuclear–mitochondrial interaction. We invite the plant research community to use and enhance this database as a shared resource to take the field in new directions.
METHODS
Cell Culture Growth, Mitochondrial Isolation, and Purity Assessment
An Arabidopsis thaliana (Landsberg erecta) cell culture was maintained in Murashige and Skoog (1962) basal medium supplemented with 3% (w/v) sucrose, 0.5 mg/L naphthaleneacetic acid, and 0.05 mg/L kinetin. Cultures were incubated in the dark at 22°C in an orbital shaker at 150 rpm. At 6 to 7 days, each flask (120 mL of cell culture) containing 8 to 10 g (fresh weight) of cells was approximately in the middle of the log-phase growth. A total of 1.0 to 1.2 L of 7-day-old dark-grown cell suspension cultures was filtered through gauze to remove medium, and the cells were dispersed in 200 mL of grinding medium (0.45 M mannitol, 50 mM sodium pyrophosphate, 0.5% [w/v] BSA, 0.5% [w/v] polyvinylpyrrolidone 40, 2 mM EGTA, and 20 mM Cys, pH 8.0). The cells were disrupted in a Waring blender by three successive 15-s bursts. Cell homogenates were filtered through cheesecloth, and mitochondria were purified by differential centrifugation on two Percoll gradients according to Millar et al. (2001a). Purified mitochondrial aliquots (500 μg) were stored at −80°C. Marker enzyme analysis for cytochrome c oxidase, alkaline pyrophosphatase, catalase, and alcohol dehydrogenase was conducted according to Millar et al. (2001a).
Based on the specific activities of marker enzymes in the mitochondria-containing fractions throughout the purification process, we concluded that plastidic contamination was decreased by 40-fold and peroxisomal contamination by 110-fold during the purification. Thus, even if the protein content of the crude pellet fraction was mainly one or the other of these two contaminating organelles (i.e., 90% plastid or peroxisomes), they could only present a maximum of 2.5% (90%/40-fold) and 1% (90%/110-fold) protein mass contamination, respectively, in our purified mitochondria. We also calculated an approximate protein mass ratio of the three organelles in the crude organelle pellet to further refine the level of contamination by peroxisomes and plastids. We did this using the protein amount found in the crude organelle pellet (∼20 mg per preparation), 70% recovery rates, and the protein amount in the purified mitochondrial fractions (∼4 mg per preparation) and peroxisome fractions (∼1.5 mg per preparation). This suggests that 28% of the crude pellet protein is mitochondrial and ∼11% is peroxisomal. We also calculated the amount of mitochondrial protein in the crude organelle pellet using cytochrome c oxidase–specific activities in the crude organelle pellet and the purified mitochondrial samples; this suggests ∼40% mitochondrial protein in the crude pellet. On average, these two methods give protein mass ratios of 60% plastidic, 30% mitochondrial, and 10% peroxisomal protein in the crude organelle pellet. Thus, the likely contamination on a protein basis in our purified mitochondrial samples is ∼1.5% plastidic in origin (60%/40-fold, with alkaline pyrophosphatase as a marker) and less than ∼0.2% peroxisomal in origin (10%/110-fold, with catalase as a marker). There was no detectible cytosolic contamination (alcohol dehydrogenase as a marker) (Millar et al., 2001b).
Sample Preparation and Mass Spectrometry
For liquid chromatography–tandem mass spectrometry (LC-MS/MS), mitochondrial samples of ∼500 μg were acetone precipitated at −20°C overnight. The resulting precipitated protein pellet was resuspended in 100 mM Tris-HCl, pH 8.5. The protein lysate was digested overnight at 37°C with trypsin 1:10 (w/w), and insoluble material was removed by centrifugation. Samples of 15 to 20 μg of digested protein were analyzed on a QStar Pulsar MS/MS system (Applied Biosystems, Foster City, CA) using an in-line Agilent 1100 capillary LC system incorporating a 0.3 × 150 mm Zorbax C18 reverse-phase column (Agilent, Palo Alto, CA) for peptide separations. Peptides were analyzed by MS over a 10-h elution period with increasing acetonitrile concentrations from 2 to 80% (v/v) in water and 0.1% (v/v) formic acid at 4 μL/min. Ions were selected automatically for the N2 collision cell using the Analyst QS software package (Applied Biosystems). Ions were selected for collision-induced distortion (CID) from an initial time-of-flight scan (1 s) if they occurred between 400 and 1500 atomic mass units (mass-to-charge ratio), had a charge series of MH2+, MH3+, or MH4+, and had an ion count >10 cps. CID spectra were accumulated for 5 s. The resulting MS/MS-derived spectra were analyzed against an in-house database comprising The Institute for Genomic Research (TIGR) and National Center for Biotechnology Information (NCBI) Arabidopsis proteins sets compiled with one allowable tryptic missed cleavage. Searches were conducted at error tolerances of ±0.15 for MS and ±0.05 for MS/MS, and only the top CID match for each spectrum was accepted.
The Pro ID matching procedure initially uses the calculated parent mass of an ion to screen the database for theoretical peptides that fall within the MS error tolerance range. A peak list composed of 20 of the most intense masses from each half of the experimentally derived CID is used to match theoretical y and b series from the candidate peptides at the specified MS/MS mass tolerance and scored. The same procedure also is undertaken for a complementary list derived from the experimental masses and the calculated parent mass. The resulting score for each CID is a combination of the number of y and b ions matched and the statistical distance this match is from a random match. The final confidence score is applied to the matched protein and is represented as a percentage value. Using this procedure, five independent mitochondrial isolations were digested and analyzed by LC-MS/MS. Proteins qualified for inclusion in the experimental set if the protein matched with a Pro ID confidence score of 98% or greater and if the protein was identified in two or more experiments. Blue-native PAGE analysis was performed according to published methods (Jansch et al., 1996). Protein spots from blue-native PAGE gels were excised and prepared for analysis by MS according to Sweetlove et al. (2002). The MS/MS collision data derived from samples were analyzed with the Pro ID MS/MS analysis component of Analyst QS as described above. Where resulting matches were <98%, manual interpretations of the spectra were undertaken to confirm the protein match. These new gel matches are incorporated in the supplemental data online along with published identifications from gel-based experiments.
Database Implementation and Data Structure
The Arabidopsis Mitochondrial Protein DataBase (AMPDB) was constructed using the MySQL database server (version 3.23; http://www.mysql.com/) and interfaces through custom-built World Wide Web pages using Hypertext Preprocessor forms version 4 (http://www.php.net/). The nonredundant nuclear protein data set used to populate the database was obtained from TIGR (http://www.tigr.org/) as the file ATH1.pep, release 3 (including supplementary data), comprising 27,288 nonredundant proteins. Arabidopsis mitochondrial (117) and chloroplast (87) open reading frame sets were obtained from NCBI (http://www.ncbi.nlm.nih.gov/). The AMPDB contains a total of 27,492 proteins. Primary attributes for proteins were produced using in-house scripts calculating molecular weight, grand average of hydropathicity (Kyte and Doolittle, 1982), and isoelectric point (Bjellqvist et al., 1993, 1994). Estimations of EST numbers for each chromosomal locus were obtained from The Arabidopsis Information Resource (TAIR; February 2003). Functional assignments were made using annotations associated with each protein entry and through homology-based comparisons with the SWISS-PROT protein database using Basic Local Alignment Search Tool (BLAST; Altschul et al., 1990).
Predictions of subcellular localization were undertaken using TargetP version 1.01 (Emanuelsson et al., 2000) with “no cut-off” set and the “Plant” option selected, Predotar version 0.5 (http://www.inra.fr/predotar/), MitoProt II version 1.0a4 (Claros and Vincens, 1996) with a discriminant function for mitochondrial proteins cutoff between 0.7 and 1.0, iPSORT (Bannai et al., 2002) with the “Plant Protein” option selected, and SubLoc (Hua and Sun, 2001) with “eukaryotic” selected. Targeting predictions were performed on the TIGR set described above. Arabidopsis mitochondrial sequence putative orthologs to yeast, human, and Rickettsia prowazekii proteins were determined using the program INPARANOID for two-way best pair-wise matches (Remm et al., 2001). The yeast mitochondrial protein set was obtained from Schon (2001) and comprises 552 proteins, the human mitochondrial protein set (615 proteins) was obtained as supplementary material from work described by Taylor et al. (2003b), and the R. prowazekii Madrid E protein set comprising 834 entries was downloaded from the Comprehensive Microbial Resource at TIGR. This database will be made publicly available at http://www.mitoz.bcs.uwa.edu.au.
Upon request, materials integral to the findings presented in this publication will be made available in a timely manner to all investigators on similar terms for noncommercial research purposes. To obtain materials, please contact A. Harvey Millar, hmillar@cyllene.uwa.edu.au.
Supplementary Material
Acknowledgments
Patrick Finnegan (University of Western Australia), Karam Singh (Commonwealth Scientific and Industrial Research Organization Plant Industry, Australia), and Steven Smith (University of Edinburgh, United Kingdom) are thanked for critical reading of the manuscript during its preparation. A.H.M. is an Australian Research Council QEII Research Fellow, and A.G. was a recipient of a Grains Research and Development Corporation Honours Scholarship. Grants from the Australian Research Council Discovery Program to D.A.D., J.W., and A.H.M. are gratefully acknowledged.
Article, publication date, and citation information can be found at www.plantcell.org/cgi/doi/10.1105/tpc.016055.
Footnotes
Online version contains Web-only data.
References
- Adams, K.L., Daley, D.O., Qiu, Y.L., Whelan, J., and Palmer, J.D. (2000). Repeated, recent and diverse transfers of a mitochondrial gene to the nucleus in flowering plants. Nature 408, 354–357. [DOI] [PubMed] [Google Scholar]
- Adams, K.L., Daley, D.O., Whelan, J., and Palmer, J.D. (2002). Genes for two mitochondrial ribosomal proteins in flowering plants are derived from their chloroplast or cytosolic counterparts. Plant Cell 14, 931–943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Altschul, S.F., Gish, W., Miller, W., Myers, E.W., and Lipman, D.J. (1990). Basic local alignment search tools. J. Mol. Biol. 215, 403–410. [DOI] [PubMed] [Google Scholar]
- Andersson, S.G., Zomorodipour, A., Andersson, J.O., Sicheritz-Ponten, T., Alsmark, U.C., Podowski, R.M., Naslund, A.K., Eriksson, A.S., Winkler, H.H., and Kurland, C.G. (1998). The genome sequence of Rickettsia prowazekii and the origin of mitochondria. Nature 396, 133–140. [DOI] [PubMed] [Google Scholar]
- Babiychuk, E., Fuangthong, M., Van Montagu, M., Inze, D., and Kushnir, S. (1997). Efficient gene tagging in Arabidopsis thaliana using a gene trap approach. Proc. Natl. Acad. Sci. USA 94, 12722–12727. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, J., and Leaver, C.J. (2001). The PET1-CMS mitochondrial mutation in sunflower is associated with premature programmed cell death and cytochrome c release. Plant Cell 13, 1803–1818. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Balk, J., Leaver, C.J., and McCabe, P.F. (1999). Translocation of cytochrome c from the mitochondria to the cytosol occurs during heat-induced programmed cell death in cucumber plants. FEBS Lett. 463, 151–154. [DOI] [PubMed] [Google Scholar]
- Bannai, H., Tamada, Y., Maruyama, O., Nakai, K., and Miyano, S. (2002). Extensive feature detection of N-terminal protein sorting signals. Bioinformatics 18, 298–305. [DOI] [PubMed] [Google Scholar]
- Bardel, J., Louwagie, M., Jaquinod, M., Jourdain, A., Luche, S., Rabilloud, T., Macherel, D., Garin, J., and Bourguignon, J. (2002). A survey of the plant mitochondrial proteome in relation to development. Proteomics 2, 880–898. [DOI] [PubMed] [Google Scholar]
- Bartoli, C., Pastori, G., and Foyer, C. (2000). Ascorbate biosynthesis in mitochondria is linked to the electron transport chain between complexes III and IV. Plant Physiol. 123, 335–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bentolila, S., Alfonso, A.A., and Hanson, M.R. (2002). A pentatricopeptide repeat-containing gene restores fertility to cytoplasmic male-sterile plants. Proc. Natl. Acad. Sci. USA 99, 10887–10892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bisanz, C., Begot, L., Carol, P., Perez, P., Bligny, M., Pesey, H., Gallois, J.L., Lerbs-Mache, S., and Mache, R. (2003). The Arabidopsis nuclear DAL gene encodes a chloroplast protein which is required for the maturation of the plastid ribosomal RNAs and is essential for chloroplast differentiation. Plant Mol. Biol. 51, 651–663. [DOI] [PubMed] [Google Scholar]
- Bjellqvist, B., Basse, B., Olsen, E., and Celis, J.E. (1994). Reference points for comparisons of two-dimensional maps of proteins from different human cell types defined in a pH scale where isoelectric points correlate with polypeptide compositions. Electrophoresis 15, 529–539. [DOI] [PubMed] [Google Scholar]
- Bjellqvist, B., Hughes, G.J., Pasquali, C., Paquet, N., Ravier, F., Sanchez, J.C., Frutiger, S., and Hochstrasser, D. (1993). The focusing positions of polypeptides in immobilized pH gradients can be predicted from their amino acid sequences. Electrophoresis 14, 1023–1031. [DOI] [PubMed] [Google Scholar]
- Braun, H.P., and Schmitz, U.K. (1999). The protein-import apparatus of plant mitochondria. Planta 209, 267–274. [DOI] [PubMed] [Google Scholar]
- Brennicke, A., Marchfelder, A., and Binder, S. (1999). RNA editing. FEMS Microbiol. Rev. 23, 297–316. [DOI] [PubMed] [Google Scholar]
- Bykova, N.V., Egsgaard, H., and Møller, I.M. (2003. a). Identification of 14 new phosphoproteins involved in important plant mitochondrial processes. FEBS Lett. 540, 141–146. [DOI] [PubMed] [Google Scholar]
- Bykova, N.V., Stensballe, A., Egsgaard, H., Jensen, O.N., and Møller, I.M. (2003. b). Phosphorylation of formate dehydrogenase in potato tuber mitochondria. J. Biol. Chem. 24, 26021–26030. [DOI] [PubMed] [Google Scholar]
- Camougrand, N., Pelissier, P., Velours, G., and Guerin, M. (1995). NCA2, a second nuclear gene required for the control of mitochondrial synthesis of subunits 6 and 8 of ATP synthase in Saccharomyces cerevisiae. J. Mol. Biol. 247, 588–596. [DOI] [PubMed] [Google Scholar]
- Chatterjee, M., Sparvoli, S., Edmunds, C., Garosi, P., Findlay, K., and Martin, C. (1996). DAG, a gene required for chloroplast differentiation and palisade development in Antirrhinum majus. EMBO J. 15, 4194–4207. [PMC free article] [PubMed] [Google Scholar]
- Chew, O., Whelan, J., and Millar, A.H. (2003). Molecular definition of the ascorbate-glutathione cycle in Arabidopsis mitochondria reveals dual-targeting of antioxidant defenses in plants. J. Biol. Chem. 278, 46869–46877. [DOI] [PubMed] [Google Scholar]
- Claros, M.G., and Vincens, P. (1996). Computational method to predict mitochondrially imported proteins and their targeting sequences. Eur. J. Biochem. 241, 779–786. [DOI] [PubMed] [Google Scholar]
- Dutilleul, C., Garmier, M., Noctor, G., Mathieu, C., Chetrit, P., Foyer, C.H., and de Paepe, R. (2003). Leaf mitochondria modulate whole cell redox homeostasis, set antioxidant capacity, and determine stress resistance through altered signaling and diurnal regulation. Plant Cell 15, 1212–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elo, A., Lyznik, A., Gonzalez, D.O., Kachman, S.D., and MacKensie, S.A. (2003). Nuclear genes encoding mitochondrial proteins for DNA and RNA metabolism are clustered in the Arabidopsis genome. Plant Cell 15, 1619–1631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Emanuelsson, O., Nielsen, H., Brunak, S., and von Heijne, G. (2000). Predicting subcellular localization of proteins based on their N-terminal amino acid sequence. J. Mol. Biol. 300, 1005–1016. [DOI] [PubMed] [Google Scholar]
- Feng, Z.-P. (2001). An overview on predicting the subcellular location of a protein. In Silico Biol. 2, 0027. [PubMed] [Google Scholar]
- Ferro, M., Salvi, D., Brugiere, S., Miras, S., Kowalski, S., Louwagie, M., Garin, J., Joyard, J., and Rolland, N. (2003). Proteomics of the chloroplast envelope membranes from Arabidopsis thaliana. Mol. Cell. Proteomics 2, 325–345. [DOI] [PubMed] [Google Scholar]
- Frerman, F.E. (1987). Reaction of electron-transfer flavoprotein ubiquinone oxidoreductase with the mitochondrial respiratory chain. Biochim. Biophys. Acta 893, 161–169. [DOI] [PubMed] [Google Scholar]
- Froehlich, J.E., Wilkerson, C.G., Ray, W.K., McAndrew, R.S., Osteryoung, K.W., Gage, D.A., and Phinney, B.S. (2003). Proteomic study of the Arabidopsis thaliana chloroplastic envelope membrane utilizing alternatives to traditional two-dimensional electrophoresis. J. Protein Res. 2, 413–425. [DOI] [PubMed] [Google Scholar]
- Fukao, Y., Hayashi, M., and Nishimura, M. (2002). Proteomic analysis of leaf peroxisomal proteins in greening cotyledons of Arabidopsis thaliana. Plant Cell Physiol. 43, 689–696. [DOI] [PubMed] [Google Scholar]
- Giegé, P., Heazlewood, J.L., Roessner-Tunali, U., Millar, A.H., Fernie, A.R., Leaver, C.J., and Sweetlove, L.J. (2003). Enzymes of glycolysis are functionally associated with the mitochondrion in Arabidopsis cells. Plant Cell 15, 2140–2151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray, M.W., Burger, G., and Lang, B.F. (1999). Mitochondrial evolution. Science 283, 1476–1481. [DOI] [PubMed] [Google Scholar]
- Gray, M.W., Burger, G., and Lang, B.F. (2001). The origin and early evolution of mitochondria. Genome Biol. 2, REVIEWS 1018.1011–1018.1015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gueguen, V., Macherel, D., Jaquinod, M., Douce, R., and Bourguignon, J. (2000). Fatty acid and lipoic acid biosynthesis in higher plant mitochondria. J. Biol. Chem. 275, 5016–5025. [DOI] [PubMed] [Google Scholar]
- Guzelin, E., Rep, M., and Grivell, L.A. (1996). Afg3p, a mitochondrial ATP-dependent metalloprotease, is involved in degradation of mitochondrially-encoded Cox1, Cox3, Cob, Su6, Su8 and Su9 subunits of the inner membrane complexes III, IV and V. FEBS Lett. 381, 42–46. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., Howell, K.A., and Millar, A.H. (2003. a). Mitochondrial complex I from Arabidopsis and rice: Orthologs of mammalian and fungal components coupled with plant-specific subunits. Biochim. Biophys. Acta 1604, 159–169. [DOI] [PubMed] [Google Scholar]
- Heazlewood, J.L., Howell, K.A., Whelan, J., and Millar, A.H. (2003. b). Towards the analysis of the rice mitochondrial proteome. Plant Physiol. 132, 230–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heazlewood, J.L., Whelan, J., and Millar, A.H. (2003. c). The products of the mitochondrial orf25 and orfB genes are FO components in the plant F1FO ATP synthase. FEBS Lett. 540, 201–205. [DOI] [PubMed] [Google Scholar]
- Herald, V.L., Heazlewood, J.L., Day, D.A., and Millar, A.H. (2003). Proteomic identification of divalent metal cation binding proteins in plant mitochondria. FEBS Lett. 537, 96–100. [DOI] [PubMed] [Google Scholar]
- Hua, S., and Sun, Z. (2001). Support vector machine approach for protein subcellular localization prediction. Bioinformatics 17, 721–728. [DOI] [PubMed] [Google Scholar]
- Jansch, L., Kruft, V., Schmitz, U.K., and Braun, H.P. (1996). New insights into the composition, molecular mass and stoichiometry of the protein complexes of plant mitochondria. Plant J. 9, 357–368. [DOI] [PubMed] [Google Scholar]
- Karlberg, O., Canback, B., Kurland, C.G., and Andersson, S.G. (2000). The dual origin of the yeast mitochondrial proteome. Yeast 17, 170–187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kispal, G., Csere, P., Guiard, B., and Lill, R. (1997). The ABC transporter Atm1p is required for mitochondrial iron homeostasis. FEBS Lett. 418, 346–350. [DOI] [PubMed] [Google Scholar]
- Kispal, G., Csere, P., Prohl, C., and Lill, R. (1999). The mitochondrial proteins Atm1p and Nfs1p are essential for biogenesis of cytosolic Fe/S proteins. EMBO J. 18, 3981–3989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kranz, R., Lill, R., Goldman, B., Bonnard, G., and Merchant, S. (1998). Molecular mechanisms of cytochrome c biogenesis: Three distinct systems. Mol. Microbiol. 29, 383–396. [DOI] [PubMed] [Google Scholar]
- Kruft, V., Eubel, H., Jansch, L., Werhahn, W., and Braun, H.P. (2001). Proteomic approach to identify novel mitochondrial proteins in Arabidopsis. Plant Physiol. 127, 1694–1710. [PMC free article] [PubMed] [Google Scholar]
- Kumar, A., et al. (2002). Subcellular localization of the yeast proteome. Genes Dev. 16, 707–719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kushnir, S., Babiychuk, E., Storozhenko, S., Davey, M.W., Papenbrock, J., De Rycke, R., Engler, G., Stephan, U.W., Lange, H., Kispal, G., Lill, R., and Van Montagu, M. (2001). A mutation of the mitochondrial ABC transporter Sta1 leads to dwarfism and chlorosis in the Arabidopsis mutant starik. Plant Cell 13, 89–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyte, J., and Doolittle, R. (1982). A simple method for displaying the hydropathic character of a protein. J. Mol. Biol. 157, 105–132. [DOI] [PubMed] [Google Scholar]
- Laloi, M. (1999). Plant mitochondrial carriers: An overview. Cell. Mol. Life Sci. 56, 918–944. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lawand, S., Dorne, A.J., Long, D., Coupland, G., Mache, R., and Carol, P. (2002). Arabidopsis A BOUT DE SOUFFLE, which is homologous with mammalian carnitine acyl carrier, is required for postembryonic growth in the light. Plant Cell 14, 2161–2173. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lister, R., Murcha, M.W., and Whelan, J. (2003). The Mitochondrial Protein Import Machinery of Plants (MPIMP) database. Nucleic Acids Res. 31, 325–327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez, M.F., Kristal, B.S., Chernokalskaya, E., Lazarev, A., Shestopalov, A.I., Bogdanova, A., and Robinson, M. (2000). High-throughput profiling of the mitochondrial proteome using affinity fractionation and automation. Electrophoresis 21, 3427–3440. [DOI] [PubMed] [Google Scholar]
- Marcotte, E.M., Xenarios, I., van der Bliek, A.M., and Eisenberg, D. (2000). Localizing proteins in the cell from their phylogenetic profiles. Proc. Natl. Acad. Sci. USA 97, 12115–12120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.H., and Heazlewood, J.L. (2003). Genomic and proteomic analysis of mitochondrial carrier proteins in Arabidopsis. Plant Physiol. 131, 443–453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Millar, A.H., Liddell, A., and Leaver, C.J. (2001a). Isolation and subfractionation of mitochondria from plants. In Methods in Cell Biology, L.A. Pon and E.A. Schon, eds (New York: Academic Press), pp. 53–74. [DOI] [PubMed]
- Millar, A.H., Sweetlove, L.J., Giege, P., and Leaver, C.J. (2001. b). Analysis of the Arabidopsis mitochondrial proteome. Plant Physiol. 127, 1711–1727. [PMC free article] [PubMed] [Google Scholar]
- Møller, I.M. (2001). Plant mitochondria and oxidative stress: Electron transport, NADPH turnover, and metabolism of reactive oxygen species. Annu. Rev. Plant Physiol. Plant Mol. Biol. 52, 561–591. [DOI] [PubMed] [Google Scholar]
- Murashige, T., and Skoog, F. (1962). A revised medium for rapid growth and bioassays with tobacco tissue culture. Physiol. Plant. 15, 473.–497. [Google Scholar]
- Nakai, K., and Horton, P. (1999). PSORT: A program for detecting sorting signals in proteins and predicting their subcellular localization. Trends Biochem. Sci. 24, 34–36. [DOI] [PubMed] [Google Scholar]
- Nijtmans, L.G., Artal Sanz, M., Bucko, M., Farhoud, M.H., Feenstra, M., Hakkaart, G.A., Zeviani, M., and Grivell, L.A. (2001). Shy1p occurs in a high molecular weight complex and is required for efficient assembly of cytochrome c oxidase in yeast. FEBS Lett. 498, 46–51. [DOI] [PubMed] [Google Scholar]
- Peeters, N., and Small, I. (2001). Dual targeting to mitochondria and chloroplasts. Biochim. Biophys. Acta 1541, 54–63. [DOI] [PubMed] [Google Scholar]
- Peltier, J.B., Emanuelsson, O., Kalume, D.E., Ytterberg, J., Friso, G., Rudella, A., Liberles, D.A., Soderberg, L., Roepstorff, P., von Heijne, G., and van Wijk, K.J. (2002). Central functions of the lumenal and peripheral thylakoid proteome of Arabidopsis determined by experimentation and genome-wide prediction. Plant Cell 14, 211–236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rabilloud, T., Kieffer, S., Procaccio, V., Louwagie, M., Courchesne, P.L., Patterson, S.D., Martinez, P., Garin, J., and Lunardi, J. (1998). Two-dimensional electrophoresis of human placental mitochondria and protein identification by mass spectrometry: Toward a human mitochondrial proteome. Electrophoresis 19, 1006–1014. [DOI] [PubMed] [Google Scholar]
- Rebeille, F., Macherel, D., Mouillon, J.M., Garin, J., and Douce, R. (1997). Folate biosynthesis in higher plants: Purification and molecular cloning of a bifunctional 6-hydroxymethyl-7,8-dihydropterin pyrophosphokinase/7,8-dihydropteroate synthase localized in mitochondria. EMBO J. 16, 947–957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remm, M., Storm, C.E.V., and Sonnhammer, E.L.L. (2001). Automatic clustering of orthologs and in-paralogs from pairwise species comparisons. J. Mol. Biol. 314, 1041–1052. [DOI] [PubMed] [Google Scholar]
- Richly, E., Chinnery, P.F., and Leister, D. (2003). Evolutionary diversification of mitochondrial proteomes: Implications for human disease. Trends Genet. 19, 356–362. [DOI] [PubMed] [Google Scholar]
- Scheffler, N.K., Miller, S.W., Carroll, A.K., Anderson, C., Davis, R.E., Ghosh, S.S., and Gibson, B.W. (2001). Two-dimensional electrophoresis and mass spectrometric identification of mitochondrial proteins from an SH-SY5Y neuroblastoma cell line. Mitochondrion 1, 161–179. [DOI] [PubMed] [Google Scholar]
- Schon, E.A. (2001). Gene products present in mitochondria of yeast and animal cells. In Mitochondria, L. Wilson and P. Matsudaira, eds (San Diego, CA: Academic Press), pp. 463–482.
- Schubert, M., Petersson, U.A., Haas, B.J., Funk, C., Schroder, W.P., and Kieselbach, T. (2002). Proteome map of the chloroplast lumen of Arabidopsis thaliana. J. Biol. Chem. 277, 8354–8365. [DOI] [PubMed] [Google Scholar]
- Sickmann, A., et al. (2003). The proteome of Saccharomyces cerevisiae mitochondria. Proc. Natl. Acad. Sci. USA 10.1073/pnas.2135385100. [DOI] [PMC free article] [PubMed]
- Smaili, S.S., Hsu, Y.T., Youle, R.J., and Russell, J.T. (2000). Mitochondria in Ca2+ signaling and apoptosis. J. Bioenerg. Biomembr. 32, 35–46. [DOI] [PubMed] [Google Scholar]
- Small, I.D., and Peeters, N. (2000). The PPR motif: A TPR-related motif prevalent in plant organellar proteins. Trends Biochem. Sci. 25, 46–47. [DOI] [PubMed] [Google Scholar]
- Suh, Y.A., Arnold, R.S., Lassegue, B., Shi, J., Xu, X., Sorescu, D., Chung, A.B., Griendling, K.K., and Lambeth, J.D. (1999). Cell transformation by the superoxide-generating oxidase Mox1. Nature 401, 79–82. [DOI] [PubMed] [Google Scholar]
- Sweetlove, L.J., Heazlewood, J.L., Herald, V., Holtzapffel, R., Day, D.A., Leaver, C.J., and Millar, A.H. (2002). The impact of oxidative stress on Arabidopsis mitochondria. Plant J. 32, 891–904. [DOI] [PubMed] [Google Scholar]
- Taylor, C.F., et al. (2003. a). A systematic approach to modeling, capturing, and disseminating proteomics experimental data. Nat. Biotechnol. 21, 247–254. [DOI] [PubMed] [Google Scholar]
- Taylor, S.W., Fahy, E., Zhang, B., Glenn, G.M., Warnock, D.E., Wiley, S., Murphy, A.N., Gaucher, S.P., Capaldi, R.A., Gibson, B.W., and Ghosh, S.S. (2003. b). Characterization of the human heart mitochondrial proteome. Nat. Biotechnol. 21, 281–286. [DOI] [PubMed] [Google Scholar]
- Werhahn, W., and Braun, H.P. (2002). Biochemical dissection of the mitochondrial proteome from Arabidopsis thaliana by three-dimensional gel electrophoresis. Electrophoresis 23, 640–646. [DOI] [PubMed] [Google Scholar]
- Werhahn, W., Jansch, L., and Braun, H.-P. (2003). Identification of novel subunits of the TOM complex from Arabidopsis thaliana. Plant Physiol. Biochem. 41, 407–416. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.